The formation and propagation of human Robertsonian chromosomes
- PMID: 40993387
- PMCID: PMC12657243
- DOI: 10.1038/s41586-025-09540-8
The formation and propagation of human Robertsonian chromosomes
Abstract
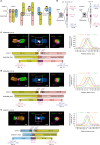
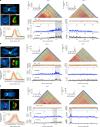
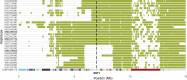
Robertsonian chromosomes are a type of variant chromosome that is commonly found in nature. Present in 1 in 800 humans, these chromosomes can underlie infertility, trisomies and increased cancer incidence1-5. They have been recognized cytogenetically for more than a century6, yet their origins have remained unknown. Here we describe complete assemblies of three human Robertsonian chromosomes. We identified a common breakpoint in SST1, a macrosatellite DNA located on chromosomes 13, 14 and 21, which commonly undergo Robertsonian translocation. SST1 is contained within a larger shared homology domain7 that is inverted on chromosome 14, which enables a meiotic crossover event that fuses the long arms of two chromosomes. Robertsonian chromosomes have two centromeric DNA arrays and have lost all ribosomal DNA. In two cases, we find that only one of the two centromeric arrays is active. In the third case, both arrays can be active but owing to their proximity, they are often encompassed by a single outer kinetochore. Thus a combination of array proximity and epigenetic changes in centromeres facilitates the stable propagation of Robertsonian chromosomes. Investigation of the assembled genomes of chimpanzee and bonobo highlights that the inversion on chromosome 14 is unique to the human genome. Resolving the structural and epigenetic features of human Robertsonian chromosomes at a molecular level provides a foundation for a broader understanding of the molecular mechanisms of structural variation and chromosome evolution.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








Update of
-
The formation and propagation of human Robertsonian chromosomes.bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614821. doi: 10.1101/2024.09.24.614821. bioRxiv. 2024. Update in: Nature. 2025 Nov;647(8091):952-961. doi: 10.1038/s41586-025-09540-8. PMID: 39386535 Free PMC article. Updated. Preprint.
References
-
- Hamerton, J. L., Canning, N., Ray, M. & Smith, S. A cytogenetic survey of 14,069 newborn infants. I. Incidence of chromosome abnormalities. Clin. Genet.8, 223–243 (1975). - PubMed
-
- Nielsen, J. & Wohlert, M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum. Genet.87, 81–83 (1991). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

