Proteomic profiling identifies miR-423-5p as a modulator of oncogenic metabolism in HCC
- PMID: 40993657
- PMCID: PMC12462032
- DOI: 10.1186/s12967-025-07039-4
Proteomic profiling identifies miR-423-5p as a modulator of oncogenic metabolism in HCC
Abstract
Background: Hepatocellular carcinoma (HCC) remains a significant clinical challenge due to limited diagnostic and therapeutic options. Non-coding RNAs (ncRNAs), such as microRNAs (miRNAs), play key roles in cancer biology. Our previous findings showed that miR-423-5p enhances anti-cancer effects on HCC patients treated with sorafenib by promoting autophagy. Here, we investigated the molecular mechanisms underlying miR-423-5p function through a comprehensive proteomic approach.
Methods: We generated an HCC cell line stably overexpressing miR-423-5p via lentiviral transduction. Total proteins were extracted from SNU-387 cells, enzymatically digested into peptides, and subsequently analysed by liquid chromatography-tandem mass spectrometry (LC-MS/M). Raw spectral data were processed and quantified using MaxQuant. Differentially expressed proteins (DEPs) were defined based on fold-change (|log2FC| ≥ 1) and false discovery rate (FDR < 0.05). The full proteomic dataset is available via the ProteomeXchange repository (identifier: PXD064869). Functional enrichment analysis of DEPs were performed using DAVID and Reactome. To assess clinical relevance, predicted and validated miR-423-5p targets were integrated with The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) dataset using GEPIA platform. Survival analyses were performed using the Kaplan-Meier method.
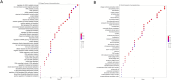
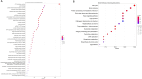
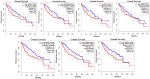
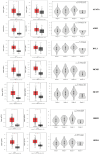
Results: Proteomic profiling identified 698 DEPs in miR-423-5p-overexpressing cells compared to controls with significant enrichment in metabolic pathways, related to purine/pyrimidine metabolism and gluconeogenesis. Integration with bioinformatic predictions and miRTarBase validation identified 43 DEPs as potential direct targets of miR-423-5p. Among these, seven proteins (ACACA, ANKRD52, DVL3, MCM5, MCM7, RRM2, SPNS1, and SRM) were significantly associated with patient prognosis in the TCGA-LIHC cohort. These targets were downregulated in miR-423-5p-overexpressing cells but upregulated in advanced-stage HCC tissues, suggesting a potential role for miR-423-5p in the regulation of HCC pathogenesis. Stage-specific expression analysis showed increased levels from stage I to III, followed by a decline at stage IV. Notably, we experimentally confirmed miR-423-5p-mediated suppression of MCM7, DVL3, IMPDH1, and SRM (SPEE), supporting their functional involvement in HCC progression.
Conclusion: Overall, our findings support a tumour-suppressive role for miR-423-5p in HCC, mediated by modulation of metabolic pathways and suppression of oncogenic proteins. These results suggest that miR-423-5p and its downstream effectors may serve as promising biomarkers and potential therapeutic targets in HCC.
Highlights: miR-423-5p acts as a tumor suppressor in HCC by targeting key nodes of pro-tumorigenic signalling. miR-423-5p significantly altered metabolic pathways, including purine/pyrimidine metabolism and gluconeogenesis. Seven miR-423-5p targets correlate with poor prognosis in TCGA-LIHC patients and are downregulated in miR-423-5p overexpressing HCC cells. miR-423-5p over-expression induces a significant downregulation of MCM7, DVL3, IMPDH1, SPEE in HCC cell models. miR-423-5p limits tumor metabolic plasticity, suggesting therapeutic potential.
Keywords: Gluconeogenesis; Hepatocellular carcinoma; Nucleotide metabolism; Overall survival; Proteomics; Spermidine synthase; Stage plot; miR-423-5p.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Competing interests: All authors declare no conflicts of interest.
Figures



References
-
- Global Burden of Disease Liver Cancer Collaboration, Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and National level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–91. - PMC - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. - PubMed
-
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

