Biomanufacturing Potential of Streamlined Cells
- PMID: 40996772
- PMCID: PMC12599491
- DOI: 10.1002/bit.70070
Biomanufacturing Potential of Streamlined Cells
Abstract
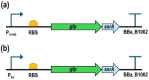
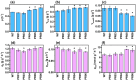
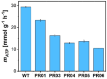
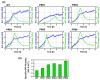
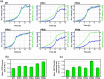
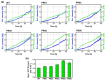
A series of Escherichia coli streamlined strains was developed by removing the expression of genes encoding extracellular structures and unessential enzymes. The streamlined strains exhibited improved metabolic performance, including lower overflow metabolism and ATP maintenance coefficient, as well as a higher growth rate, compared to their parental strain. The intracellular levels of ATP were monitored using a genetic sensor, showing the improved resource stewardship of the streamlined cells. The streamlined strains were tested as cell factories to produce plasmid DNA (pDNA) in batch cultures, exhibiting a 23% increase in the specific pDNA production rate, compared to the parental strain. Recombinant protein expression was evaluated in microbioreactors in batch and fed-batch modes. In batch mode, recombinant protein yield from biomass was up to 82% higher in the streamlined strains than in the parental strain. Furthermore, in fed-batch mode, the recombinant protein yield was 79% greater in the streamlined cells compared to the parental strain. Our results show the benefits of reducing cellular complexity on the biomanufacturing of pDNA and recombinant proteins in culture schemes typical of industrial settings.
Keywords: ATP maintenance; microbial engineering; minimal cells; proteome reduction.
© 2025 The Author(s). Biotechnology and Bioengineering published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

