The multiomics blueprint of the individual with the most extreme lifespan
- PMID: 40997805
- PMCID: PMC12629823
- DOI: 10.1016/j.xcrm.2025.102368
The multiomics blueprint of the individual with the most extreme lifespan
Abstract
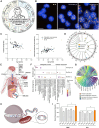
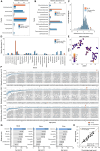
Extreme human lifespan, exemplified by supercentenarians, presents a paradox in understanding aging: despite advanced age, they maintain relatively good health. To investigate this duality, we have performed a high-throughput multiomics study of the world's oldest living person, interrogating her genome, transcriptome, metabolome, proteome, microbiome, and epigenome, comparing the results with larger matched cohorts. The emerging picture highlights different pathways attributed to each process: the record-breaking advanced age is manifested by telomere attrition, abnormal B cell population, and clonal hematopoiesis, whereas absence of typical age-associated diseases is associated with rare European-population genetic variants, low inflammation levels, a rejuvenated bacteriome, and a younger epigenome. These findings provide a fresh look at human aging biology, suggesting biomarkers for healthy aging, and potential strategies to increase life expectancy. The extrapolation of our results to the general population will require larger cohorts and longitudinal prospective studies to design potential anti-aging interventions.
Keywords: aging; epigenetics; genetics; microbiome; supercentenarian.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.E. declares past grants from Ferrer International and Incyte and personal fees from Quimatryx and Eucerin, outside of the submitted work.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

