Cryo-EM structure of the full-length LGR4-RSPOs complex and a targeting nanobody for anti-obesity therapy
- PMID: 40998774
- PMCID: PMC12462478
- DOI: 10.1038/s41467-025-63410-5
Cryo-EM structure of the full-length LGR4-RSPOs complex and a targeting nanobody for anti-obesity therapy
Abstract
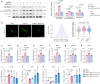
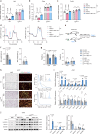
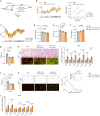
Obesity poses a substantial threat to human health but lacks effective management. Recent advancements in large-scale deep sequencing and cryo-electron microscopy (cryo-EM) have transformed drug discovery paradigms. Leveraging prior genetics investigation, we pinpointed Leucine-rich repeat-containing G protein-coupled receptor 4 (LGR4) as a promising target for combating obesity. Here, we present cryo-EM structures of full-length LGR4 alone and in complex with RSPO2(FU). Notably, we develop an inhibitory nanobody (NB21) that blocks the binding of RSPO1/2 to LGR4, and we also determine the structure of the LGR4-NB21 complex. NB21-mFc (NB21 fused with mouse IgG2) effectively inhibits the canonical Wnt signaling pathway, thereby enhancing mitochondrial respiration and thermogenesis in beige adipocytes. In vivo, NB21-mFc increases energy expenditure by promoting the browning of white fat, conferring resistance to both diet-induced and genetic (ob/ob) obesity. Furthermore, LGR4 deficiency abolishes the effects of NB21-mFc in boosting the browning program and subsequent weight reduction. In summary, our study unveils structural insights into the LGR4-RSPOs and LGR4-NB21 complexes, paving the way for the development of an LGR4-targeting nanobody for weight loss.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.W. and G.N. are listed as inventors on a patent application entitled “LGR4-targeting nanobody NB21 and use thereof in anti-obesity treatment” (PCT/CN2025/076673; CN120058937), filed by Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The patent covers the NB21 antibody and its therapeutic use in obesity. The remaining authors declare no competing interests.
Figures



References
-
- Bjerregaard, L. G. & Baker, J. L. Change in overweight from childhood to early adulthood and risk of type 2 diabetes. N. Engl. J. Med.378, 2537–2538 (2018). - PubMed
-
- Calle, E. E., Rodriguez, C., Walker-Thurmond, K. & Thun, M. J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med.348, 1625–1638 (2003). - PubMed
-
- Gregg, E. W. & Shaw, J. E. Global health effects of overweight and obesity. N. Engl. J. Med.377, 80–81 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

