The genetic diversity of Indonesian cattle has been shaped by multiple introductions and adaptive introgression
- PMID: 40998788
- PMCID: PMC12462482
- DOI: 10.1038/s41467-025-62692-z
The genetic diversity of Indonesian cattle has been shaped by multiple introductions and adaptive introgression
Abstract
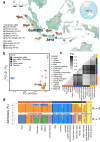
Genetic diversity is a crucial resource in livestock, determining their traits and ability to respond to selection. Indonesian cattle are unique due to their history of admixture involving both zebu (Bos indicus) and banteng (B. javanicus), and may therefore contain novel cattle genetic resources. We generated whole genome sequences from 126 Indonesian cattle, 51 domesticated banteng and three captive banteng. We show that Indonesian cattle have very high genetic diversity, especially the Madura breed due to introgression from banteng and possibly other Bos species, contributing up to 36.6% of the Madura's genome. We find that Indonesian zebu ancestry can be traced to at least three distinct ancestral populations, two of which were introduced more than 1345 years ago from mainland Southeast or eastern Asia. Peaks and valleys in banteng ancestry across the genome in admixed breeds suggest that both negative and positive selection act on introgressed haplotypes. Despite adaptive introgression being mainly breed-specific, we found evidence that some phenotypes, such as coat color, have experienced convergent adaptive introgression. Overall, our results provide insights into the historical movement of cattle in Asia, and showcase the potential for genetic improvement of cattle by identifying ~3.5 million novel SNPs introgressed into Indonesian cattle.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- The State of the World’s Biodiversity for Food and Agriculture: FAO COMMISSION ON GENETIC RESOURCES FOR FOOD AND AGRICULTURE ASSESSMENTS •. Food and Agriculture Organization of the United Nations Food & Agriculture Org (2019).
MeSH terms
Grants and funding
- DFF8049-00098B/Det Frie Forskningsråd (Danish Council for Independent Research)
- Starting Grant 853442/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- CF20-0355/Carlsbergfondet (Carlsberg Foundation)
LinkOut - more resources
Full Text Sources

