Oncogenic driver mutations underlie the spatial tumour immune landscape of non-small cell lung cancer
- PMID: 40998804
- PMCID: PMC12462439
- DOI: 10.1038/s41467-025-63465-4
Oncogenic driver mutations underlie the spatial tumour immune landscape of non-small cell lung cancer
Abstract
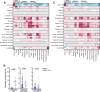
Lung adenocarcinoma (LUAD) is a molecularly diverse form of lung cancer characterized by distinct oncogenic driver mutations that influence both tumour biology and clinical outcomes. Understanding the interplay between these oncogenic drivers and the tumour microenvironment (TME) is crucial for improving therapeutic strategies and patient management. Here, we investigate the impact of driver mutations on the composition and spatial architecture of the TME in LUAD. Using imaging mass cytometry (IMC), we analyse tumour samples from 157 LUAD patients, integrating genomic and clinical data to link specific mutations with tumour characteristics. Unique patterns are associated with mutated KRAS and EGFR tumours with TP53 co-mutations, suggesting these co-mutations reshape the TME and promote resistance to tyrosine kinase inhibitors (TKIs). Overall, our findings highlight the complex interplay between oncogenic driver mutations and the TME in LUAD, underscoring the importance of integrating genomic and cellular data to understand the underlying tumour behaviour and prognosis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Imyanitov, E. N., Iyevleva, A. G. & Levchenko, E. V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol. /Hematol.157, 103194 (2021). - PubMed
-
- Del Re, M. et al. Concise review: resistance to Tyrosine Kinase inhibitors in non-small cell lung cancer: the role of cancer stem cells. Stem Cells36, 633–640 (2018). - PubMed
-
- Lim, S. M., Syn, N. L., Cho, B. C. & Soo, R. A. Acquired resistance to EGFR targeted therapy in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Treat. Rev.65, 1–10 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

