EMB is essential for enteric nervous system development mediated by PI3K signaling
- PMID: 40999499
- PMCID: PMC12465749
- DOI: 10.1186/s13073-025-01538-1
EMB is essential for enteric nervous system development mediated by PI3K signaling
Abstract
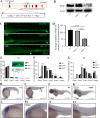
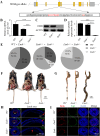
Background: The enteric nervous system (ENS), which arises from enteric neural crest cells (ENCCs), plays important roles in many aspects of gastrointestinal tract function, including motility, secretions, blood flow and hormone release. Defects in ENS development could lead to a broad range of disorders, including Hirschsprung's disease (HSCR), which is characterized by missing nerve cells in the distal segment of the colon. Here, we identify EMB as an evolutionarily conserved regulator of ENS development.
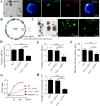
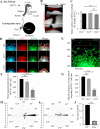
Methods: We first examined EMB expression in human and mouse intestines using scRNA-seq data and immunofluorescence staining. To investigate its role in ENS development, we constructed Emb-knockout zebrafish and mouse models. To explore the underlying mechanisms, we focused on ENCCs and analyzed their proliferation and migration using migration assays in explant guts and organoid cultures. Finally, we assessed rare EMB variants in a cohort of HSCR patients.
Results: In zebrafish, loss of emb leads to a decrease number of enteric neurons and impaired intestinal transit ability. In mice, knockout of Emb causes HSCR-like phenotypes and defects. In vitro experiments, including explant mouse gut and organoid cultures, show that EMB is required for both the proliferation and migration of ENCCs. Mechanistically, EMB binds to and recruits the phosphatase complex PP2A to the cellular membrane to facilitate the activation of PI3K-AKT pathway, thereby promoting ENCCs development. Indeed, application of PI3K or AKT agonists partially restores the ENS developmental defects in zebrafish emb mutants. Furthermore, rare variants of EMB may potentially contribute to the pathology of HSCR in humans.
Conclusions: EMB is required for ENS development by regulating the proliferation and migration of the ENCCs. Mechanistically, EMB recruits PP2A to the cell membrane, reducing cytoplasmic dephosphorylation activity and promoting the activation of the PI3K signaling pathway.
Keywords: EMB; Enteric nervous system; Hirschsprung’s Disease; Neural crest cell; PI3K pathway.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All protocols involving human sample collection in this study were approved by the Ethics Committee of Tongji Medical College of Huazhong University of Science and Technology (Ethical approval number: 2020-S226, 2021-S033). All relevant parts of the research were conducted in accordance with the Declaration of Helsinki, and written informed consent was obtained from all participants or their legal guardians. Experiments about zebrafish or mice were approved by the Scientific Ethics Committee of Huazhong Agricultural University (Ethical approval number: HZAUMO-2017-048). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Heuckeroth RO. Hirschsprung disease - integrating basic science and clinical medicine to improve outcomes. Nat Rev Gastroenterol Hepatol. 2018;15(3):152–67. - PubMed
-
- Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9(5):286–94. - PubMed
-
- Huang T, Hou Y, Wang X, Wang L, Yi C, Wang C, et al. Direct Interaction of Sox10 With Cadherin-19 Mediates Early Sacral Neural Crest Cell Migration: Implications for Enteric Nervous System Development Defects. Gastroenterology. 2022;162(1):179-92.e11. - PubMed
-
- Zhou B, Feng C, Sun S, Chen X, Zhuansun D, Wang D, et al. Identification of signaling pathways that specify a subset of migrating enteric neural crest cells at the wavefront in mouse embryos. Dev Cell. 2024;59(13):1689-706.e8. - PubMed
-
- Landman KA, Simpson MJ, Newgreen DF. Mathematical and experimental insights into the development of the enteric nervous system and Hirschsprung’s disease. Dev Growth Differ. 2007;49(4):277–86. - PubMed
MeSH terms
Substances
Grants and funding
- 2023Y9159/Joint Funds for the innovation of Science and Technology, Fujian province
- 81721005/Innovative Research Group Project of the National Natural Science Foundation of China
- 82071685/National Natural Science Foundation of China
- 2019YBKY026/Clinical Research Pilot Project of Tongji Hospital
- 2020BCB008/Hubei Provincial Key Research and Development Program
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

