This is a preprint.
Next-Generation Multiplexed Targeted Proteomics Quantifies Post-Translational Modifications, Compound-Protein Interactions, and Disease Biomarkers with High Throughput
- PMID: 41000625
- PMCID: PMC12458933
- DOI: 10.1101/2025.09.10.675380
Next-Generation Multiplexed Targeted Proteomics Quantifies Post-Translational Modifications, Compound-Protein Interactions, and Disease Biomarkers with High Throughput
Abstract
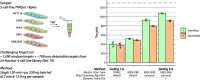
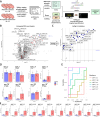
The GoDig platform enables sensitive, multiplexed targeted pathway proteomics without manual scheduling or synthetic standards. Here we present GoDig 2.0, which increases sample multiplexing to 35-fold, improves time efficiency and reduces scan delays for higher success rates, and allows flexible spectral and elution library generation from different mass spectrometry data types. GoDig 2.0 measures 2.4× more targets than GoDig 1.0, quantifying >99% of 800 peptides in a single run. We compiled a library of 23,989 human phosphorylation sites from a phosphoproteomic dataset and used it to profile kinase signaling differences across cell lines. In human brain tissue, we established a hyperphosphorylated tau assay including pTau127, revealing potential biomarkers for Alzheimer's disease. We also quantified diglycyl-lysine peptides to assess polyubiquitin branching. Finally, we built a library of 20,946 reactive cysteines and profiled covalent compound-protein interactions spanning diverse pathways. GoDig 2.0 enables high-throughput analyses of site-specific protein modifications across many biological contexts.
Conflict of interest statement
COMPETING INTERESTS J.D.C. is a full-time employee of Thermo Fisher Scientific, which manufactures the Orbitrap Eclipse and Orbitrap Ascend mass spectrometers.
Figures




References
-
- Shuken S. R. “An Introduction to Mass Spectrometry-Based Proteomics.” J. Proteome Res. 2023, 22, 2151–2171. - PubMed
-
- Marx V. “Targeted proteomics.” Nature Methods 2013, 10, 19–22. - PubMed
-
- Mallick P.; et al. “Computational prediction of proteotypic peptides for quantitative proteomics.” Nature Biotechnol. 2007, 25, 125–131. - PubMed
-
- Pauletti B. A.; et al. “Typic: A Practical and Robust Tool to Rank Proteotypic Peptides for Targeted Proteomics.” J. Proteome Res. 2023, 22, 539–545. - PubMed
-
- Yu Q.; et al. “Sample multiplexing-based targeted pathway proteomics with real-time analytics reveals the impact of genetic variation on protein expression.” Nature Comm. 2023, 14, 555.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
