This is a preprint.
Pore-Based RNA Evaluation for Control of Integrity, Sequence, and Errors - Quality Control (PRECISE-QC)
- PMID: 41000762
- PMCID: PMC12458368
- DOI: 10.1101/2025.09.20.677417
Pore-Based RNA Evaluation for Control of Integrity, Sequence, and Errors - Quality Control (PRECISE-QC)
Abstract
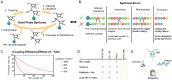
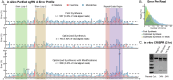
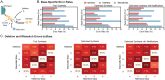
RNA is at the forefront of therapeutics and gene editing technologies. Yet, RNA synthesis remains expensive and low-yield. Consequently, most oligo manufacturers abstain from synthesizing RNA oligos longer than 60-mers. Solid-phase synthesis is the current standard production method but is often fraught with low coupling yields for canonical nucleotides and even poorer coupling for modifications. This results in high levels of byproducts such as truncations and RNA infidelity. Existing analytical methods can only provide quality control metrics such as RNA length distribution or limited composition information for short oligos. Here, we developed a standard quality control metric using Oxford Nanopore direct RNA sequencing to obtain direct insight into RNA length distribution, sequence, and presence of RNA modification sites. Our pipeline identifies error-prone regions and truncation sites that occur during synthesis. Furthermore, problematic steps in the synthesis are identified and repaired. We show that our platform can produce and assess CRISPR guide RNAs with high-fidelity and higher cleavage activity, and further, that modifications can be reliably detected. We envision that our tool will serve as an integral method for quality control pipelines that assess the integrity and accuracy of synthetic RNAs and guide the improved synthesis and yield of synthesized RNAs.
Keywords: Nanopore Sequencing; Quality Control; RNA Modifications; RNA synthesis; Solid phase synthesis.
Conflict of interest statement
Conflict of Interest The authors declare no competing interests.
Figures


References
-
- Flemmich L, Bereiter R, Micura R. Chemical synthesis of modified RNA. Angew Chem Int Ed Engl. 2024;63: e202403063. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
