This is a preprint.
A cross model spatial and single-cell atlas reveals the conserved involvement of osteopontin in polycystic kidney disease
- PMID: 41000888
- PMCID: PMC12458445
- DOI: 10.1101/2025.09.17.676846
A cross model spatial and single-cell atlas reveals the conserved involvement of osteopontin in polycystic kidney disease
Abstract
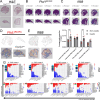
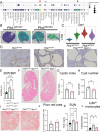
Polycystic kidney disease (PKD) arises from mutations in cilia-associated genes, such as PKD1 and PKD2, expressed in renal epithelial cells, leading to progressive kidney dysfunction and end-stage kidney disease (ESKD). PKD patients exhibit significant heterogeneity in disease progression, largely due to genetic and environmental modifiers. Like patients, mouse models of PKD also exhibit significant heterogeneity with regards to the gene mutated, age of disease onset, and rate of disease progression. To elucidate the cellular and molecular consequences of these variables, we constructed an integrated single-cell and spatial transcriptomics atlas across mouse models of PKD, mapping changes in cell type composition, gene expression, and intercellular signaling networks across the whole atlas and within individual models. Consistently across models, single cell RNA sequencing (scRNAseq) data revealed increased Spp1 (osteopontin) expression and signaling from PKD-enriched clusters to Ly6clo monocytes. Global deletion of Spp1 in Pkd1 RC/RC mice resulted in reduced cyst severity, improved kidney function, and reduced Ly6clo monocyte numbers, suggesting that SPP1 signaling to Ly6clo monocytes promotes PKD progression. We also created a freely available, searchable website (https://bmblx.bmi.osumc.edu/scPKD/) that can be used to identify cross- and intra-model specific changes in gene expression, guiding researchers to new therapeutic targets for treating PKD.
Keywords: Polycystic kidney disease; macrophages; osteopontin; single cell RNA sequencing; spatial transcriptomics.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
