This is a preprint.
Complex Genetics and Regulatory Drivers of Hypermobile Ehlers-Danlos Syndrome: Insights from Genome-Wide Association Study Meta-analysis
- PMID: 41001447
- PMCID: PMC12458961
- DOI: 10.1101/2025.09.19.25336146
Complex Genetics and Regulatory Drivers of Hypermobile Ehlers-Danlos Syndrome: Insights from Genome-Wide Association Study Meta-analysis
Abstract
Background: Hypermobile Ehlers-Danlos syndrome (hEDS) is the most common subtype of EDS, a group of heritable connective tissue disorders. Clinically, hEDS is defined by generalized joint hypermobility and chronic musculoskeletal pain, but its impact extends beyond the musculoskeletal system. Affected individuals frequently experience autonomic, gastrointestinal, immune, and neuropsychiatric involvement, highlighting both the multisystemic nature of the condition and challenges of diagnosis. In contrast to other EDS subtypes with defined genetic causes, the molecular basis of hEDS has remained elusive.
Methods: We conducted a genome-wide association study (GWAS) of hEDS across three case controls studies, including 1,815 cases and 5,008 ancestry-matched controls. Fixed-effects meta-analysis of 6.2 million variants was complemented with LDAK gene-based association testing, transcriptome-wide association studies, and integrative annotation across multiple tissues and cell types including eQTLs, enhancer marks and open chromatin accessibility profiles, supported by luciferase assays on one candidate variant. LD-score genetic correlations were assessed between hEDS and 19 frequently reported comorbid conditions.
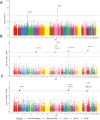
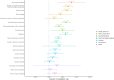
Results: Two loci reached genome-wide significance, including a regulatory region near the atypical chemokine receptor 3 gene (ACKR3) on chromosome 2. Functional annotation supports ACKR3 risk alleles colocalize with eQTLs in tibial nerve, alter enhancer activity, and generate a de novo AHR transcription factor regulatory site, implicating neuroimmune and pain signaling pathways. Gene-based and transcriptome-wide analyses identified common variants in a locus containing multiple candidates, including SLC39A13, a zinc transporter critical for connective tissue development previously implicated in a rare form of EDS, and PSMC3, a gene involved in central nervous system development. LD-score regression revealed significant genetic correlations between hEDS and joint hypermobility, myalgic encephalomyelitis/chronic fatigue syndrome, fibromyalgia, depression, anxiety, autism spectrum disorder, migraine, and gastrointestinal diseases.
Conclusions: These results establish the first evidence of common variant contributions to hEDS, supporting a complex, multisystem model involving neuroimmune-stromal dysregulation. Our findings add novel indications to hEDS pathogenesis and provide solid foundations for future molecular definition and therapeutic discovery.
Keywords: Genome-wide association study; Hypermobile Ehlers-Danlos Syndrome; neurodevelopment; neuroimmune signaling.
Figures


References
-
- Demmler J.C. et al. Diagnosed prevalence of Ehlers-Danlos syndrome and hypermobility spectrum disorder in Wales, UK: a national electronic cohort study and case-control comparison. BMJ Open 9, e031365 (2019).
-
- Daylor V. et al. Defining the Chronic Complexities of hEDS and HSD: A Global Survey of Diagnostic Challenges, Life-Long Comorbidities, and Unmet Needs. J Clin Med 14(2025).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
