This is a preprint.
Germline Variants Influence Chronic Liver Disease Progression through Distinct Pathways
- PMID: 41001506
- PMCID: PMC12458504
- DOI: 10.1101/2025.09.16.25335186
Germline Variants Influence Chronic Liver Disease Progression through Distinct Pathways
Abstract
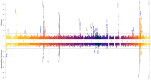
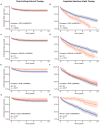
Cirrhosis and hepatocellular carcinoma (HCC) are long-term complications of chronic liver disease (CLD). In this large multi-ancestry genome-wide association study of all-cause cirrhosis (35,481 cases, 2.36M controls) and HCC (6,680 cases, 1.76M controls), we identified 27 loci associated with cirrhosis (10 novel) and 11 with HCC (three novel). Three novel cirrhosis loci were replicated in independent cohorts (e.g. FGF21, RPTOR, and IFNL3/4). Fifteen cirrhosis loci exhibited differential effects on cirrhosis risk via underlying etiologies, and six HCC loci influenced HCC risk indirectly via cirrhosis. In a gene-burden analysis of rare variants from whole-genome sequencing data in the VA Million Veteran Program (n=102,677), we identified GSTA5 as a novel cirrhosis-associated gene, while APOB and ATP9B were associated with and replicated for HCC. A high genetic risk score for cirrhosis was associated with a nearly doubled risk of CLD progressing to cirrhosis (HR=1.94, P=2×10-68) and of cirrhosis progressing to HCC (HR=1.65, P=7×10-08). Finally, among individuals with chronic hepatitis C who underwent antiviral therapy, cirrhosis risk was modified by variants in PNPLA3, IFNL3/4, and CD81 following pegylated interferon-α therapy, and by APOE lead variant following direct-acting antiviral therapy. These findings provide new insights into the complex genetic architecture of CLD progression with potential clinical and therapeutic implications.
Conflict of interest statement
D.E.K. receives funding to institution from AstraZeneca, Gilead, Bausch and Exact Sciences. J.A.L. and C.C.T. report grants from Alnylam Pharmaceuticals, Inc., AstraZeneca Pharmaceuticals LP, Biodesix, Inc, Janssen Pharmaceuticals, Inc., Novartis International AG, Parexel International Corporation through the University of Utah or Western Institute for Veteran Research outside the submitted work. The authors who are affiliated with Regeneron Genetics Center declare competing financial interests as employees. J.G. has received lecture fee from Illumina. UK Biobank and has received multiple grants from academic, charitable and industry sources outside of the submitted work. Q.M.A. reports Research Grant Funding: Coordinator of the EU IMI-2 LITMUS consortium, which is funded by the EU Horizon 2020 programme and EFPIA. AstraZeneca, Boehringer Ingelheim, Intercept. Consultancy on behalf of Newcastle University: Alimentiv, Akero, AstraZeneca, Axcella, 89Bio, Boehringer Ingelheim, Bristol Myers Squibb, Corcept, Echosens, Enyo Pharma, Galmed, Genfit, Genentech, Gilead, GlaxoSmithKline, Hanmi, HistoIndex, Intercept, Inventiva, Ionis, IQVIA, Janssen, Madrigal, Medpace, Merck, Metadeq, NGMBio, North Sea Therapeutics, Novartis, Novo Nordisk, PathAI, Pfizer, Pharmanest, Poxel, Prosciento, Resolution Therapeutics, Roche, Ridgeline Therapeutics, RTI, Shionogi, Terns. Speaker: Fishawack, Integritas Communications, Kenes, Novo Nordisk, Madrigal, Medscape, Springer Healthcare. Royalties: Elsevier Ltd. M.G.L. was supported by the Doris Duke Foundation (Award 2023-0224) and US Department of Veterans Affairs Biomedical Research and Development Award IK2-BX006551. M.G.L. reports research grants from MyOme and consulting fees from BridgeBio, unrelated to the present work. L.Y., A.O., Y.C. and E.K.S. are supported by R01DK131787 (to E.K.S.) R01DK128871 (to E.K.S.), The University of Michigan MBIOFar Award and The University of Michigan Department of Internal Medicine. D.G. is Chief Executive Officer of Sequoia Genetics, a private R&D consultancy that works with investors, pharma and biotech in using human genetic data to inform drug discovery and development. Outside of this work, DG has financial interests in several biotech companies. All other authors have no conflict of interest to declare.
Figures



References
-
- Younossi Z.M., et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016). - PubMed
-
- Kisseleva T. & Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol 18, 151–166 (2021). - PubMed
-
- Anstee Q.M., Reeves H.L., Kotsiliti E., Govaere O. & Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol 16, 411–428 (2019). - PubMed
Publication types
Grants and funding
- R01 DK131787/DK/NIDDK NIH HHS/United States
- UM1 DK126194/DK/NIDDK NIH HHS/United States
- U01 AI152960/AI/NIAID NIH HHS/United States
- OT2 OD026556/OD/NIH HHS/United States
- R01 DK131033/DK/NIDDK NIH HHS/United States
- OT2 OD026551/OD/NIH HHS/United States
- U24 OD023121/OD/NIH HHS/United States
- OT2 OD026552/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- OT2 OD025337/OD/NIH HHS/United States
- C06 RR020128/RR/NCRR NIH HHS/United States
- OT2 OD025277/OD/NIH HHS/United States
- OT2 OD026555/OD/NIH HHS/United States
- OT2 OD026550/OD/NIH HHS/United States
- OT2 OD026553/OD/NIH HHS/United States
- OT2 OD023205/OD/NIH HHS/United States
- I01 BX003362/BX/BLRD VA/United States
- WT_/Wellcome Trust/United Kingdom
- U24 OD023163/OD/NIH HHS/United States
- OT2 OD023206/OD/NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- OT2 OD026548/OD/NIH HHS/United States
- UL1 TR002529/TR/NCATS NIH HHS/United States
- U2C OD023196/OD/NIH HHS/United States
- F30 HL172382/HL/NHLBI NIH HHS/United States
- OT2 OD025315/OD/NIH HHS/United States
- R01 DK134575/DK/NIDDK NIH HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- I01 CX002010/CX/CSRD VA/United States
- OT2 OD026557/OD/NIH HHS/United States
- UL1 TR002243/TR/NCATS NIH HHS/United States
- R01 DK128871/DK/NIDDK NIH HHS/United States
- OT2 OD026554/OD/NIH HHS/United States
- IK2 BX006551/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
