Mapping the Proteomic Landscape of Pancreatic Cancer: Prognostic Insights and Subtype Stratification
- PMID: 41003647
- PMCID: PMC12548992
- DOI: 10.1158/2767-9764.CRC-25-0229
Mapping the Proteomic Landscape of Pancreatic Cancer: Prognostic Insights and Subtype Stratification
Abstract
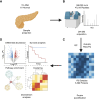
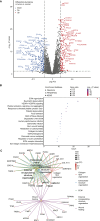
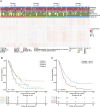
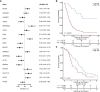
Pancreatic ductal adenocarcinoma (PDA) is an aggressive malignancy that lacks reliable biomarkers to guide treatment decisions. Effective prognostic tools are needed to improve its clinical management. We conducted a comprehensive proteomic analysis on 115 PDA patient samples with matched adjacent normal tissue. A 20-protein diagnostic panel was identified (LGALS1, ANXA2, LGALS3BP, CTSD, S100P, COL12A1, SFN, THBS2, CTHRC1, THBS1, SERPINB5, LAMC2, POSTN, CEACAM6, CTSE, PLEC, PKM, S100A11, TAGLN2, ALDOA). Consensus clustering analysis identified four prognostic proteomic subtypes. Subtypes with poorer prognoses exhibited upregulation of neutrophil degranulation, extracellular matrix remodeling, focal adhesion, Mesenchymal Epithelial Transition, collagen formation, and PI3K-Akt-mTOR-related pathways, indicating a predominance of basal-like and activated stromal features. In tumors with homologous recombination deficiency or Catalogue of Somatic Mutations in Cancer Signature-3, several immune-related proteins were enriched. An 18-protein (PURB, SDCBP2, CD2BP2, GALM, SERPINA3, OAS3, FAN1, ZPR1, KRT2, NUDT2, SMNDC1, SERPINA4, CUTA, WDR36, POSTN, CLEC11A, PEX14, and PI4KA) risk score was developed and validated using multicox regression analyses with LASSO regularization. The risk score demonstrated independent prognostic significance for overall survival and recurrence, and was validated in an independent proteomic dataset generated using a different proteomic technology. This study thus introduces four novel prognostic PDA subtypes, and an 18-protein risk score validated in an independent dataset, which shows promise for improving survival prediction and could serve as a valuable tool for personalized treatment guidance.
Significance: The findings from this study have significant implications for the future of pancreatic cancer management. By identifying a 20-protein panel with diagnostic and screening potential, this research provides a foundation for developing early detection tools for PDA, an aggressive cancer with limited treatment options. The classification of PDA into four proteomic subtypes with distinct prognostic outcomes paves the way for subtype-specific therapeutic approaches, allowing clinicians to better stratify patients based on their risk profiles. Additionally, the validated 18-protein risk score, which enhances survival prediction and operates independently of existing clinical variables, represents a promising tool for personalized prognostic assessments. Incorporating these proteomic-based biomarkers into clinical practice could improve diagnostic accuracy, guide individualized treatment decisions, and ultimately enhance patient outcomes in PDA. This study underscores the potential of proteomic profiling to improve cancer treatment by providing targeted, actionable insights into tumor biology.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
A.T. Aref, M. Pathan, Z. Noor, A. Anees, D. Bucio-Noble, E.K. Sykes, P.J Robinson and R.R. Reddel report a patent number 2025900265 pending. N. Waddell reports grants from National Health and Medical Research Council during the conduct of the study; other from genomiQa outside the submitted work; and other support from the Australian Pancreatic Genome Initiative executive (member). O. Kondrashova reports personal fees from AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Figures
References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. - PubMed
-
- Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet 2020;395:2008–20. - PubMed
-
- Ducreux M, Cuhna AS, Caramella C, Hollebecque A, Burtin P, Goéré D, et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015;26(Suppl 5):v56–68. - PubMed
-
- National Comprehensive Cancer Network . Pancreatic adenocarcinoma version 2. Plymouth Meeting (PA): National Comprehensive Cancer Network; 2023[cited 2023 Jul]. Available from:https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

