Distinct cell state ecosystems for nodular lymphocyte-predominant Hodgkin lymphoma
- PMID: 41006203
- PMCID: PMC12475200
- DOI: 10.1038/s41467-025-63339-9
Distinct cell state ecosystems for nodular lymphocyte-predominant Hodgkin lymphoma
Abstract
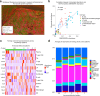
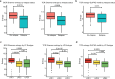
Nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) is a rare cancer, and few studies have comprehensively investigated the immune microenvironment and rare lymphocyte-predominant (LP) cells. Here we develop a NLPHL specific lymphocyte-predominant ecotype (LPE) model to identify 34 distinct cell states across 14 cell types that co-occur within 3 LPEs for 171 cases. LPE1 and LPE2 were characterized by immunosuppressive microenvironments with high expression of B2M on LP cells, CD8 T-cell exhaustion, immune checkpoint genes expressed by follicular T-cells, and an improved freedom from progression compared to LPE3 in training (n = 109, with 65% LPE1/2) and validation cohorts (n = 62, with 61% LPE1/2). We validate the co-occurrence and co-localization of cell states using spatial transcriptomics. Protein expression of HLA-I and HLA-II on LP cells and SSTR2 on dendritic cells was predictive of LPE1 (C-statistic=0.69), LPE2 (C-statistic=0.79), and LPE3 (C-statistic=0.60). This study establishes a clinically relevant biologic categorization for NLPHL.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.M. reports consulting and honoraria with Lilly, Bristol-Myers Squibb, Abbvie, and Roche. M.D. reports research funding from AstraZeneca, Genentech, Varian Medical Systems and Illumina, ownership interest in CiberMed and Foresight Diagnostics, consultancy from AstraZeneca, Boehringer Ingelheim, BMS, Genentech, Gritstone Oncology, Illumina, Regeneron and Roche, and multiple issued and pending patents including patents licensed to Foresight Diagnostics and Roche. A.A.A. reports consultancy for Celgene, Chugai, Genentech, Gilead, Janssen, Pharmacyclics and Roche, scientific advisory board membership in the Lymphoma Research Foundation, and Professional Affiliations with the American Society of Hematology, American Society of Clinical Oncology, American Society of Clinical Investigation, Leukemia & Lymphoma Society, Research Funding from the National Cancer Institute, the National Heart, Lung, and Blood Institute, the US National Institutes of Health, Celgene, BMS and Pfizer, patent filings including patent issued, licensed and with royalties paid from FortySeven, a patent pending and Licensed to Foresight, a patent pending relating to MARIA, a patent issued and licensed to CiberMed, a patent issued and a patent pending to CiberMed, patents issued to Idiotype Vaccines, and a patent issued, licensed and with royalties paid from Roche, and equity ownership interests in CiberMed Inc., ForeSight Diagnostics, FortySeven Inc. and CARGO Therapeutics. The remaining authors declare no competing interests.
Figures





References
-
- Regula, D. P., Hoppe, R. T. & Weiss, L. M. Nodular and diffuse types of lymphocyte predominance Hodgkin’s disease. N. Engl. J. Med318, 214–219 (1988). - PubMed
-
- Eichenauer, D. A. et al. Long-term follow-up of patients with nodular lymphocyte-predominant Hodgkin lymphoma treated in the HD7 to HD15 trials: a report from the German Hodgkin Study Group. J. Clin. Oncol.10.1200/JCO.19.00986 (2019). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

