A protein-proximity screen reveals Ebola virus co-opts the mRNA decapping complex through the scaffold protein EDC4
- PMID: 41006235
- PMCID: PMC12475165
- DOI: 10.1038/s41467-025-63392-4
A protein-proximity screen reveals Ebola virus co-opts the mRNA decapping complex through the scaffold protein EDC4
Abstract
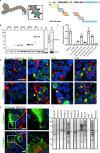
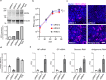
The interaction of host and Ebola virus (EBOV) proteins is required for establishing infection. In this study, we use proximity-dependent biotinylation to identify cellular proteins that bind to EBOV proteins encoded by six of the seven viral genes. Hits are computationally mapped onto a human protein-protein interactome and annotated with viral proteins, confirming known EBOV-host protein interactions and revealing previously undescribed interactions and processes. This approach efficiently arranges proteins into functional complexes associated with single viral proteins. Focused characterization of interactions between EBOV VP35 and the mRNA decapping complex shows that VP35 binds the scaffold protein EDC4 through the C-terminal subdomain, with both proteins colocalizing in EBOV-infected cells. siRNA depletion of EDC4, DCP2, and EDC3 reduces virus replication by inhibiting early viral RNA synthesis. Overall, the analytical approach efficiently identifies EBOV protein interactions with cellular protein complexes, providing a deeper understanding of replication mechanisms for therapeutic intervention.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Update of
-
A protein-proximity screen reveals Ebola virus co-opts the mRNA decapping complex through the scaffold protein EDC4.Res Sq [Preprint]. 2024 Feb 2:rs.3.rs-3838220. doi: 10.21203/rs.3.rs-3838220/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Sep 26;16(1):8485. doi: 10.1038/s41467-025-63392-4. PMID: 38352529 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- P01 AI120943/AI/NIAID NIH HHS/United States
- R01 AI114814/AI/NIAID NIH HHS/United States
- R01AI114814/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P01AI120943/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Medical

