Decoding structural transitions from CdSe nanoclusters to quantum dots through dynamic nuclear polarization NMR
- PMID: 41006241
- PMCID: PMC12475265
- DOI: 10.1038/s41467-025-62724-8
Decoding structural transitions from CdSe nanoclusters to quantum dots through dynamic nuclear polarization NMR
Abstract
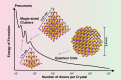
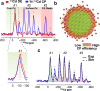
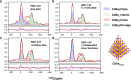
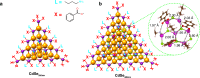
Achieving control over quantum dot size is essential for their performance, but remains challenging due to the limited atomic-level understanding of quantum dot growth. In this study, we employ signal-enhancing Dynamic Nuclear Polarization solid-state NMR to investigate the structural features of intermediate CdSe clusters and mature quantum dots to gain valuable insights. By integrating quantum mechanical calculations with experimental data, we leverage 113Cd chemical shift to elucidate local Cd environments at various positions, decoding the complex ligand distribution on cluster surfaces. Our findings reveal that ligand distribution is stabilizing through inter-ligand hydrogen bonds, while minimizing steric clashes during ligand packing on space-constrained planar facets. This study underscores the unique capability of 113Cd NMR to probe local Cd environments, offering a framework for monitoring structural transitions and improving size control during quantum dot growth.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: R.A.F. is a cofounder and consultant for Schrodinger Inc. The remaining authors declare no competing interests.
Figures




References
-
- Steigerwald, M. L. & Brus, L. E. Semiconductor crystallites: a class of large molecules. Acc. Chem. Res.23, 183–188 (1990). - DOI
-
- Dai, X., Deng, Y., Peng, X. & Jin, Y. Quantum-dot light-emitting diodes for large-area displays: towards the dawn of commercialization. Adv. Mater. 29, 1607022 (2017). - PubMed
-
- Nomura, M. & Arakawa, Y. Light sources: Shaking quantum dots. Nat. Photonics6, 9–10 (2012). - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

