Gtf2i-encoded transcription factor Tfii-i regulates myelination via Sox10 and Mbp regulatory elements
- PMID: 41006289
- PMCID: PMC12474925
- DOI: 10.1038/s41467-025-63500-4
Gtf2i-encoded transcription factor Tfii-i regulates myelination via Sox10 and Mbp regulatory elements
Abstract
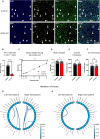
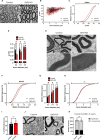
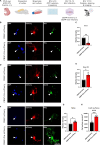
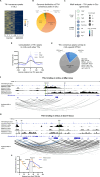
The transcriptional regulatory network governing the differentiation and functionality of oligodendrocytes (OLs) is essential for the formation and maintenance of the myelin sheath, and hence for the proper function of the nervous system. Perturbations in the intricate interplay of transcriptional effectors within this network can lead to a variety of nervous system pathologies. In this study, we identify Gtf2i-encoded general transcription factor II-I (Tfii-i) as a regulator of key myelination-related genes. Gtf2i deletion from myelinating glial cells in male mice leads to functional alterations in central nervous system (CNS) myelin, including elevated mRNA and protein expression levels of myelin basic protein (Mbp), the central myelin component, enhanced connectivity properties, and thicker myelin wrapping axons with increased diameters. These changes resulted in faster axonal conduction across the corpus callosum (CC), and improved motor coordination. Furthermore, we show that in mature OLs (mOLs), Tfii-i directly binds to regulatory elements of Sox10 and Mbp. In the peripheral nervous system (PNS), Gtf2i deletion from Schwann cells (SCs) leads to hypermyelination of the tibial branch of the sciatic nerve (SN). These findings add to our understanding of myelination regulation and specifically elucidate a cell-autonomous mechanism for Tfii-i in myelinating glia transcriptional network.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

