Optimization of DNA Fragmentation Techniques to Maximize Coverage Uniformity of Clinically Relevant Genes Using Whole Genome Sequencing
- PMID: 41008666
- PMCID: PMC12468493
- DOI: 10.3390/diagnostics15182294
Optimization of DNA Fragmentation Techniques to Maximize Coverage Uniformity of Clinically Relevant Genes Using Whole Genome Sequencing
Abstract
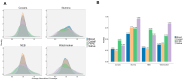
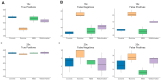
Background: Coverage uniformity is pivotal in whole genome sequencing (WGS), as uneven read distributions can obscure clinically relevant variants and compromise downstream analyses. While enzyme-based fragmentation methods for WGS library preparation are widely used, they can introduce sequence-specific biases that disproportionately affect high-GC or low-GC regions. Here, we compare four PCR-free WGS library preparation workflows-one employing mechanical fragmentation and three based on enzymatic fragmentation-to assess their impact on coverage uniformity and variant detection. Results: Libraries were generated with Coriell NA12878 and DNA isolated from DNA blood, saliva, and formalin-fixed paraffin-embedded (FFPE) samples. Sequencing was performed on an Illumina NovaSeq 6000, followed by alignment to the human reference genome (GRCh38/hg38) and local realignment. We assessed coverage at both chromosomal and gene levels, including 504 clinically relevant genes detected in the TruSight™ Oncology 500 (TSO500) panel. Additionally, we examined the relationship between GC content and normalized coverage, as well as variant detection across high- and low-GC regions. Conclusions: Our findings show that mechanical fragmentation yields a more uniform coverage profile across different sample types and across the GC spectrum. Enzymatic workflows, on the other hand, demonstrated more pronounced coverage imbalances, particularly in high-GC regions, potentially affecting the sensitivity of variant detection. This effect was evident in analyses focusing on the TSO500 gene set, where uniform coverage is critical for accurate identification of disease-associated variants and for minimizing false negatives. Downsampling experiments further revealed that mechanical fragmentation maintained lower Single Nucleotide Polymorphism (SNPs) false-negative and false-positive rates at reduced sequencing depths, thereby highlighting the advantages of consistent coverage for resource-efficient WGS. This study introduces a novel framework for evaluating WGS coverage uniformity, providing guidance for optimizing library preparation protocols in clinical and translational research. By quantifying how fragmentation strategies influence coverage depth and variant calling accuracy, laboratories can refine their sequencing workflows to ensure more reliable detection of clinically actionable variants-especially in high-GC regions often implicated in hereditary disease and oncology.
Keywords: GC-bias; PCR-free library preparation; adaptive focused acoustics (AFA) fragmentation; chromosomal coverage; coverage uniformity; enzymatic fragmentation; library preparation; next-generation sequencing (NGS); variant detection; whole genome sequencing (WGS).
Conflict of interest statement
Several authors (Vanessa Process, Madan Ambavaram, Sameer Vasantgadkar, Sushant Khanal, Martina Werner, Greg Endress, Ulrich Thomann, and Eugenio Daviso) are employees of Covaris LLC., a PerkinElmer Company. This study compares DNA fragmentation methods, including the truCOVER PCR-free Library Prep Kit, which is developed and manufactured by Covaris LLC. Covaris LLC. provided funding and resources for this research. The findings indicate superior performance for mechanical fragmentation, which is the technology employed by Covaris’s truCOVER product.
Figures





References
-
- Ellingford J.M., Barton S., Bhaskar S., Williams S.G., Sergouniotis P.I., O’Sullivan J., Lamb J.A., Perveen R., Hall G., Newman W.G., et al. Whole Genome Sequencing Increases Molecular Diagnostic Yield Compared with Current Diagnostic Testing for Inherited Retinal Disease. Ophthalmology. 2016;123:1143–1150. doi: 10.1016/j.ophtha.2016.01.009. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

