The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure
- PMID: 41009471
- PMCID: PMC12469698
- DOI: 10.3390/ijms26188899
The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure
Abstract
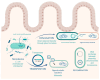
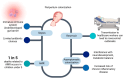
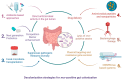
Colistin, a polymyxin antibiotic reintroduced as a last-resort therapy against multidrug-resistant Gram-negative bacteria, is increasingly being compromised by the emergence of plasmid-mediated colistin resistance genes (mcr-1 to mcr-10). The human gut microbiota serves as a major reservoir and transmission hub for these resistance determinants, even among individuals without prior colistin exposure. This review explores the mechanisms, dissemination, and clinical implications of mcr-mediated colistin resistance within the gut microbiota, highlighting its role in horizontal gene transfer, colonization, and environmental persistence. A comprehensive synthesis of the recent literature was conducted, focusing on epidemiological studies, molecular mechanisms, neonatal implications and decolonization strategies. The intestinal tract supports the enrichment and exchange of mcr genes among commensal and pathogenic bacteria, especially under antibiotic pressure. Colistin use in agriculture has amplified gut colonization with resistant strains in both animals and humans. Surveillance gaps remain, particularly in neonatal populations, where colonization may occur early and persist silently. Promising interventions, such as fecal microbiota transplantation and phage therapies, are under investigation but lack large-scale clinical validation. The gut microbiome plays a central role in the global spread of colistin resistance. Mitigating this threat requires integrated One Health responses, improved diagnostics for gut colonization, and investment in microbiome-based therapies. A proactive, multisectoral approach is essential to safeguard colistin efficacy and address the expanding threat of mcr-mediated resistance.
Keywords: antimicrobial resistance (AMR); colistin resistance; gut microbiota; horizontal gene transfer (HGT); mcr genes; neonatal colonization; plasmid-mediated resistance.
Conflict of interest statement
The authors declare no conflicts of interest. Regional surveillance emphasis reflects available data and may not capture all global trends.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

