Molecular Epidemiology of SARS-CoV-2 Detected from Different Areas of the Kandy District of Sri Lanka from November 2020-March 2022
- PMID: 41012617
- PMCID: PMC12474409
- DOI: 10.3390/v17091189
Molecular Epidemiology of SARS-CoV-2 Detected from Different Areas of the Kandy District of Sri Lanka from November 2020-March 2022
Abstract
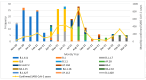
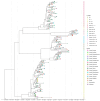
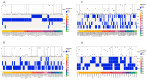
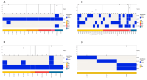
A comprehensive analysis of the molecular epidemiology of SARS-CoV-2 in the Kandy District of Sri Lanka from November 2020 to March 2022 was conducted to address the limited genomic surveillance data available across the country. The study investigated the circulating SARS-CoV-2 lineages, their temporal dynamics, and the associated mutational profiles in the study area. A total of 280 SARS-CoV-2-positive samples were selected, and 252 complete genomes were successfully sequenced using Oxford Nanopore Technology. Lineage classification was performed using the EPI2ME tool, while phylogenetic relationships were inferred through maximum likelihood and time-scaled phylogenetic trees using IQ-TREE2 and BEAST, respectively. Amino acid substitutions were analyzed to understand lineage-specific mutation patterns. Fifteen SARS-CoV-2 lineages were identified, and of those B.1.411 (36%) was the most prevalent, followed by Q.8 (21%), AY.28 (9.5%), and the Delta and Omicron variants. The lineage distribution showed a temporal shift from B.1.411 to Alpha, Delta, and finally the Omicron, mirroring the global trends. Time to the most recent common ancestor analyses provided estimates for the introduction of major variants, while mutation analysis revealed the widespread occurrence of D614G in the spike protein and lineage-specific mutations across structural, non-structural, and accessory proteins.Detection of the Epsilon variant (absent in other national-level studies) in November 2020, highlighted the regional heterogeneity viral spread. This study emphasizes the importance of localized genomic surveillance to capture the true diversity and evolution of SARS-CoV-2, to facilitate containment strategies in resource-limited settings.
Keywords: Kandy district; SARS-CoV-2 infection; Sri Lanka; mutation profile; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Van Der Made C.I., Simons A., Schuurs-Hoeijmakers J., Van Den Heuvel G., Mantere T., Kersten S., Van Deuren R.C., Steehouwer M., Van Reijmersdal S.V., Jaeger M., et al. Presence of Genetic Variants among Young Men with Severe COVID-19. JAMA-J. Am. Med. Assoc. 2020;324:663–673. doi: 10.1001/jama.2020.13719. - DOI - PMC - PubMed
-
- Kumar R., Srivastava Y., Muthuramalingam P., Singh S.K., Verma G., Tiwari S., Tandel N., Beura S.K., Panigrahi A.R., Maji S., et al. Understanding Mutations in Human SARS-CoV-2 Spike Glycoprotein: A Systematic Review & Meta-Analysis. Viruses. 2023;15:856. doi: 10.3390/v15040856. - DOI - PMC - PubMed
-
- Jeewandara C., Jayathilaka D., Ranasinghe D., Hsu N.S., Ariyaratne D., Jayadas T.T., Panambara Arachchige D.M., Lindsey B.B., Gomes L., Parker M.D., et al. Genomic and Epidemiological Analysis of SARS-CoV-2 Viruses in Sri Lanka. Front. Microbiol. 2021;12:722838. doi: 10.3389/fmicb.2021.722838. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

