Living Together Apart: Quantitative Perspectives on the Costs and Benefits of a Multipartite Genome Organization in Viruses
- PMID: 41012703
- PMCID: PMC12474326
- DOI: 10.3390/v17091275
Living Together Apart: Quantitative Perspectives on the Costs and Benefits of a Multipartite Genome Organization in Viruses
Abstract
Background: Multipartite viruses individually package their multiple genome segments into virus particles, necessitating the transmission of multiple virus particles for effective viral spread. This dependence poses a cost in the form of reduced transmission compared to monopartite viruses, which only have a single genome segment. The notable cost of a multipartite genome organization has spurred debate on why multipartite viruses are so common among plant viruses, including a search for benefits associated with this organizational form.
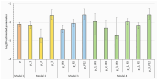
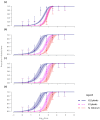
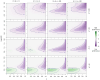
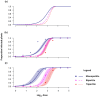
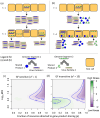
Methods: We investigated the costs and benefits of multipartite viruses with three approaches. First, we reanalyzed dose-response data to measure the cost of multipartition to between-host transmission for multipartite viruses. Second, we developed a simulation model to explore when the sharing of viral gene products between cells is beneficial. Third, we tested whether multipartite viruses have a broad host range by estimating the host range for plant viruses using metagenomics data.
Results: We find that the observed cost to transmission exceeds theoretical predictions. We predict that a virus gene-product-sharing strategy only confers benefits under limited conditions, suggesting that this strategy may not be common. Our results suggest that multipartite and segmented viruses have broader host ranges than monopartite viruses.
Conclusions: Our analyses also suggest there is limited evidence for the costs and benefits of a multipartite organization, and we argue that the diversity of multipartite virus-host systems demands pluralistic explanatory frameworks.
Keywords: dose response; evolution; gene-product sharing; host range; infection model; multipartite virus; plant virus; segmented virus.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

