Large-scale screening of genes responsible for silique length and seed size in Brassica Napus via pooled CRISPR library
- PMID: 41013219
- PMCID: PMC12465755
- DOI: 10.1186/s12864-025-11979-y
Large-scale screening of genes responsible for silique length and seed size in Brassica Napus via pooled CRISPR library
Abstract
Background: Enhancing rapeseed (Brassica napus, B. napus) yield is critical for ensuring global vegetable oil security. However, yield is heavily influenced by silique development and seed size, the enhancement of which is limited by scarce genetic resources. The CRISPR/Cas9 system has emerged as a powerful tool for constructing genome-wide mutant libraries, even in polyploid crops with complex genomes.
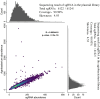
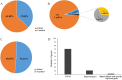
Results: The transcriptome-wide association study (TWAS) data, tissue-specific expression profiles data and reported genes were integrated to identify candidate genes regulating silique development and seed size. We constructed a sgRNA library targeting these genes and generated a CRISPR/Cas9 editing mutant library through genetic transformation. Specifically, 6124 sgRNAs were designed for 1739 candidate genes with ≦ 4 orthologues. 681 T0 plants were obtained through genetic transformation, which harbor 453 sgRNAs. Of 408 T0 plants analyzed, 151 (37.00%) exhibited successful gene editing events, targeting 84 candidate genes. Ten homozygous mutant plants were isolated and preliminary phenotypic analysis was performed in mutants targeting the BnaHRDs. The results suggest that mutations in BnaHRD.A03 and BnaHRD.C03 may modulate plant height (PH), main inflorescence length (MIL), silique length (SL), effective silique number per plant (ENS), seed number per silique (SNPS), and thousand-seed weight (TSW).
Conclusions: This study harnessed the CRISPR/Cas9 technology to establish a preliminary library of gene-edited mutants in B. napus, thereby laying a robust foundation for the future screening of candidate genes pertaining to silique development and seed size. Furthermore, this study provides a methodological framework for rapid functional gene discovery in B. napus through CRISPR-based approaches.
Keywords: Brassica Napus; CRISPR; Gene editing; Seed size; Silique length.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All authors have been informed and have given their consent. Competing interests: The authors declare no competing interests.
Figures






References
-
- Li L, Chen B, Yan G, Gao G, Xu K, Xie T, et al. Strategies and progress in the research and utilization of Chinese rapeseed germplasm resources. J Plant Genetic Resour. 2020;21:1–19. (Chinese abstract in English).
-
- Hua W, Li R, Zhan G, Liu J, Li J. Maternal control of seed oil content in Brassica napus: the role of silique wall photosynthesis. Plant J. 2011;69:432–44. - PubMed
-
- Zhou X, Dai L, Wang P, Liu Y, Xie Z, Zhang H, et al. Mining favorable alleles for five agronomic traits from the elite rapeseed cultivar zhongshuang11 by QTL mapping and integration. Crop J. 2021;9:1449–59.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

