Droplet Digital PCR-Based Detection of Clarithromycin Resistance on Rapid Urease Test Samples Predicts Helicobacter pylori Eradication Success: A New Zealand Cohort Study
- PMID: 41013956
- PMCID: PMC12475888
- DOI: 10.1111/hel.70075
Droplet Digital PCR-Based Detection of Clarithromycin Resistance on Rapid Urease Test Samples Predicts Helicobacter pylori Eradication Success: A New Zealand Cohort Study
Abstract
Background: Helicobacter pylori (H. pylori) infection is a major cause of peptic ulcer disease and gastric cancer. Rising clarithromycin resistance has significantly reduced the efficacy of standard triple therapy. In Aotearoa New Zealand, the prevalence and impact of antibiotic resistance remain incompletely defined, limiting the development of effective treatment strategies.
Methods and aims: This study evaluated the feasibility and clinical utility of detecting clarithromycin resistance genes using droplet digital polymerase chain reaction (ddPCR) on stored Rapid Urease Test (RUT) samples-a relatively novel application of molecular diagnostics. We also assessed the association between resistance status and treatment outcomes following empiric first-line triple therapy. Patients with positive RUT tests during gastroscopy were treated with omeprazole-based triple therapy and followed up with H. pylori stool antigen testing to confirm eradication.
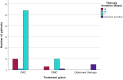
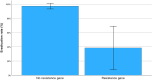
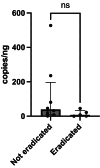
Results: Among 84 patients, clarithromycin resistance genes were detected in 13 (15.5%). Overall eradication was achieved in 74 (88.1%) patients. However, eradication success was significantly lower in those with resistance (38.5%) compared to those without (97.2%, p < 0.001). Infection burden, treatment regimen, and duration were not associated with eradication rates, supporting resistance status as the primary determinant of treatment outcome. Resistance rates were similar between Māori and Pacific patients (18.2%) and other ethnic groups (14.8%), although sample sizes limited definitive conclusions.
Conclusions: ddPCR testing on stored RUT samples is a feasible and effective method for detecting clarithromycin resistance. This study demonstrates that clarithromycin resistance, rather than infection burden, treatment regimen, or duration, drives eradication failure in New Zealand. Tailored therapy based on molecular resistance testing may enhance treatment success and support antibiotic stewardship. These findings justify the development of PCR-guided treatment pathways and provide a strong rationale for extending this approach to non-invasive stool-based testing suitable for use in primary care and screening programs.
Keywords: Helicobacter pylori; clarithromycin resistance; tailored therapy.
© 2025 The Author(s). Helicobacter published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Correa P., “Human Gastric Carcinogenesis: A Multistep and Multifactorial Process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention1,” Cancer Research 52, no. 24 (1992): 6735–6740. - PubMed
-
- Hsiang J., Selvaratnam S., Taylor S., et al., “Increasing Primary Antibiotic Resistance and Ethnic Differences in Eradication Rates of <styled-content style="fixed-case"> Helicobacter pylori </styled-content> Infection in New Zealand—A New Look at an Old Enemy,” New Zealand Medical Journal 126, no. 1384 (2013): 64–76. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

