Reference-guided assembly of metagenomes with MetaCompass
- PMID: 41015028
- PMCID: PMC12570322
- DOI: 10.1016/j.crmeth.2025.101186
Reference-guided assembly of metagenomes with MetaCompass
Abstract
Metagenomic studies have primarily relied on de novo assembly for reconstructing genes and genomes from microbial mixtures. While reference-guided approaches have been employed in the assembly of single organisms, they have not been used in a metagenomic context. Here, we develop an effective approach for reference-guided metagenomic assembly that can complement and improve upon de novo metagenomic assembly methods for certain organisms. Such approaches will be increasingly useful as more genomes are sequenced and made publicly available.
Keywords: CP: genetics; CP: microbiology; comparative assembly; metagenome assembly; metagenomics; microbiome; reference-guided assembly.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

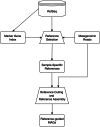
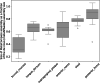
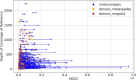
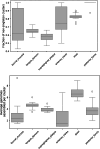

Update of
-
MetaCompass: Reference-guided Assembly of Metagenomes.ArXiv [Preprint]. 2024 Mar 3:arXiv:2403.01578v1. ArXiv. 2024. Update in: Cell Rep Methods. 2025 Oct 20;5(10):101186. doi: 10.1016/j.crmeth.2025.101186. PMID: 38903742 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

