In silico characterization of chromosomally integrated blaCTX-M genes among clinical Enterobacteriaceae in Africa: insights from whole-genome analysis
- PMID: 41019522
- PMCID: PMC12463934
- DOI: 10.3389/fmicb.2025.1655907
In silico characterization of chromosomally integrated blaCTX-M genes among clinical Enterobacteriaceae in Africa: insights from whole-genome analysis
Abstract
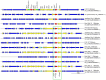
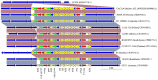
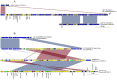
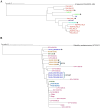
Antimicrobial resistance (AMR) mediated by extended-spectrum β-lactamases (ESBLs) is a growing global concern, particularly among Enterobacteriaceae. The CTX-M-type ESBLs, encoded by the bla CTX-M gene, are of significant public health importance due to their high prevalence and broad geographic distribution. Typically located on plasmids and often co-occurring with other AMR genes, bla CTX-M contributes to multidrug resistance (MDR). However, increasing evidence suggests secondary chromosomal integration of bla CTX-M, sometimes alongside other resistance determinants. The extent and implications of this mechanism remain poorly characterized, especially in Africa, where genomic surveillance is limited. In this study, we retrieved 295 chromosomal sequences of Enterobacteriaceae of African origin from the GenBank and performed in silico predictions of bla CTX-M and other AMR genes. bla CTX-M-carrying sequences were further characterized by in silico multilocus sequence typing and genome annotation. Chromosomal insertions were identified through alignment with reference genomes. Overall, 47 of 295 sequences (15.9%) harbored the bla CTX-M gene, with the highest prevalence in Klebsiella pneumoniae (29/157, 18.5%), followed by Escherichia coli (13/72, 18.1%), Enterobacter spp. (4/38, 10.5%), and Shigella spp. (1/12, 8.3%). The most common allele was bla CTX-M-15 (31/47, 66.0%), followed by bla CTX-M-14 (12/47, 25.5%), bla CTX-M-55 (3/47, 6.4%), and bla CTX-M-27 (1/27, 3.7%). Co-occurrence of bla CTX-M with additional AMR genes was frequently observed, with integration events often associated with mobile genetic elements such as ISEcp1 and IS26. Notably, strains from the same hospital setting were phylogenetically related and shared sequence types and AMR gene profiles, suggesting local clonal dissemination. These findings reveal a notable presence of chromosomally integrated bla CTX-M among African Enterobacteriaceae, frequently in association with other resistance genes, thereby facilitating stable MDR propagation independent of plasmid maintenance. This evolutionary adaptation may have significant implications for the persistence and spread of MDR in clinical settings.
Keywords: Africa; Enterobacteriaceae; IS26; ISEcp1; blaCTX-M; chromosomal.
Copyright © 2025 Shawa, Chambaro, Kamboyi, Sulwe, Chizimu, Nasilele, Ogata, Samutela, Zorigt, Mudenda, Simbotwe, Nsofwa, Chanda, Chabala, Nundwe, Ndebe, Sinjani, Hayashida, Nao, Chilengi, Sawa, Suzuki, Hang’ombe, Kajihara and Higashi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bassetti M., Pecori D., Sibani M., Corcione S., De Rosa F. G. (2015). Epidemiology and treatment of MDR Enterobacteriaceae. Curr. Treat. Options Infect. Dis. 7, 291–316. doi: 10.1007/s40506-015-0065-1 - DOI
-
- Beyene A. M., Gizachew M., Yousef A. E., Haileyesus H., Abdelhamid A. G., Berju A., et al. (2024). Multidrug-resistance and extended-spectrum beta-lactamase-producing lactose-fermenting Enterobacteriaceae in the human-dairy interface in Northwest Ethiopia. PLoS One 19:e0303872. doi: 10.1371/journal.pone.0303872, PMID: - DOI - PMC - PubMed
-
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

