Pan-immune system, mobilome and resistome in Streptococcus suis
- PMID: 41021386
- PMCID: PMC12479173
- DOI: 10.1099/mgen.0.001521
Pan-immune system, mobilome and resistome in Streptococcus suis
Abstract
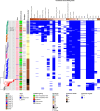
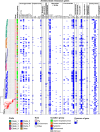
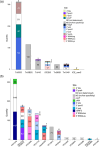
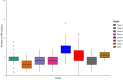
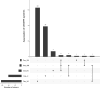
Streptococcus suis is a bacterial pathogen responsible for infections in pigs and in wild fauna that can also lead to severe infections in humans. Increasing antimicrobial resistance (AMR) has been described for this zoonotic pathogen worldwide. Since most of these AMR genes are carried by mobile genetic elements (MGEs), they can largely disseminate by horizontal gene transfer. Taking advantage of the large set of genomes available for this species, an exhaustive search of integrative and conjugative elements (ICEs) and integrative and mobilizable elements (IMEs) was undertaken in a representative set of 400 selected high-quality genomes of S. suis. We examined how these elements vary across phylogenetic clades and ecotypes and their association with AMR genes and defence systems (DSs), including restriction-modification (RM), CRISPR and also less studied DSs. This investigation identified 569 ICEs, belonging to the 7 families previously described in streptococci, inserted in 12 distinct specific integration sites. Additionally, 1,035 IMEs characterized by 11 distinct relaxase families and integrated in 10 specific chromosomal sites were detected in the 400 genomes of S. suis. New associations between ICE/IME and AMR genes were discovered. A huge diversity of putative DSs was observed including 2,035 RM systems, 124 CRISPR systems and systems belonging to 20 other categories, most of them described as efficient against phages and plasmids. Furthermore, most of the spacers associated with CRISPR systems target these MGEs rather than integrative elements. In addition, many integrative elements appear to carry an orphan methylase that could help them escape RM systems. Altogether, this points out that ICEs and IMEs are spared by DSs and play a major role in AMR dissemination in S. suis. In addition, most of the strains have the full set of genes required for competence, i.e. for the acquisition of extracellular DNA by natural transformation. This suggests a high risk of AMR dissemination in S. suis.
Keywords: CRISPR; Streptococcus suis; antimicrobial resistance genes; competence; defence systems; integrative and conjugative elements; integrative and mobilizable elements; restriction–modification systems; virulence.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
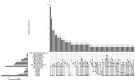

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

