Identifying genetic determinants of outer retinal function in mice using a large-scale gene-targeted screen
- PMID: 41021661
- PMCID: PMC12503315
- DOI: 10.1371/journal.pgen.1011886
Identifying genetic determinants of outer retinal function in mice using a large-scale gene-targeted screen
Abstract
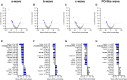
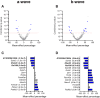
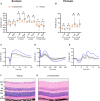
Electroretinography (ERG) provides a noninvasive functional measure of multiple cell types of the outer retina. We conducted an ERG-based screen of 530 single-gene knockout mouse strains generated as part of the International Mouse Phenotyping Consortium, representing 2.5% of all protein-coding genes, to identify genetic variants affecting retinal function. We identified 30 strains with significantly altered ERG amplitudes. Two of the genes identified, Cfap418 and Syne2, have been previously reported with outer retinal dysfunction, thereby serving as internal controls that validate our screening protocol. Of the remaining 28 genes newly associated with altered retinal function, the majority lacked a contemporaneous histopathology correlate, highlighting the importance of ERG in early detection of functional abnormalities. A rare homozygous missense variant in FCHSD2, the human orthologue of one of the 28 genes identified, was found in a patient presenting with retinal degeneration that lacked a molecular diagnosis. This report represents a useful resource for future investigations into the molecular mechanisms driving inherited retinal diseases and demonstrates the power of large-scale ERG screening in identifying novel genetic determinants of retinal function.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

