Genome-resolved biogeography of Phaeocystales, cosmopolitan bloom-forming algae
- PMID: 41022706
- PMCID: PMC12480563
- DOI: 10.1038/s41467-025-63565-1
Genome-resolved biogeography of Phaeocystales, cosmopolitan bloom-forming algae
Abstract
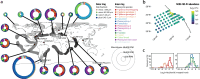
Phaeocystales, comprising the genus Phaeocystis and an uncharacterized sister lineage, are nanoplanktonic haptophytes widespread in the global ocean. Several species form mucilaginous colonies and influence key biogeochemical cycles, yet their underlying diversity and ecological strategies remain underexplored. Here, we present new genomic data from 13 strains, including three high-quality reference genomes (N50 > 30 kbp), and integrate previous metagenome-assembled genomes to resolve a robust phylogeny. Divergence timing of P. antarctica aligns with Miocene cooling and Southern Ocean isolation. Genomic traits reveal metabolic flexibility, including mixotrophic nitrogen acquisition in temperate waters and gene expansions linked to polar nutrient adaptation. Concordantly, transcriptomic comparisons between temperate and polar Phaeocystis suggest Southern Ocean populations experience iron and B12 limitation. We also identify signatures of horizontal gene transfer and endogenous giant virus/virophage insertions. Together, these findings highlight Phaeocystales as an ecologically versatile and geographically widespread lineage shaped by evolutionary innovation and adaptation to contrasting environmental stressors.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Baumann, M. E. M., Lancelot, C., Brandini, F. P., Sakshaug, E. & John, D. M. The taxonomic identity of the cosmopolitan prymnesiophyte phaeocystis: A morphological and ecophysiological approach. J. Mar. Syst.5, 5–22 (1994).
-
- Schoemann, V., Becquevort, S., Stefels, J., Rousseau, V. & Lancelot, C. Phaeocystis blooms in the global ocean and their controlling mechanisms: A review. J. Sea Res.53, 43–66 (2005).
-
- Smith, W. O. & Trimborn, S. Phaeocystis: a global enigma. Ann. Rev. Mar. Sci.16, 417–441 (2024). - PubMed
-
- Le Quéré, C. et al. Ecosystem dynamics based on plankton functional types for global ocean biogeochemistry models. Glob. Chang Biol.11, 2016–2040 (2005).
-
- Vogt, M. et al. Global marine plankton functional type biomass distributions: Phaeocystis spp. Earth Syst. Sci. Data4, 107–120 (2012).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

