A loss-of-function human ADAR variant activates innate immune response and promotes bowel inflammation
- PMID: 41022715
- PMCID: PMC12480066
- DOI: 10.1038/s41467-025-63554-4
A loss-of-function human ADAR variant activates innate immune response and promotes bowel inflammation
Abstract
Inflammatory bowel disease (IBD) arises from genetic-environmental interactions. Adenosine deaminases acting on RNA 1 (ADAR), an RNA-editing enzyme converting adenosine (A) to inosine (I), is essential for tissue homeostasis. Here we report that intestinal ADAR deficiency contributes to IBD pathogenesis in humans with reduced ADAR expression in patient intestinal crypts. Genetic or pharmacological inhibition of ADAR in mice causes spontaneous ileitis and colitis. Organoid studies show that ADAR loss leads to double-strand RNA (dsRNA) and endogenous retroviruses (ERVs) accumulation, disrupting intestinal homeostasis via melanoma differentiation-associated protein 5 (MDA5)-mediated dsRNA sensing and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling. Editome analyses identify Mda5 as an ADAR target, and edited Mda5 exhibits impaired dsRNA sensing. The human ADAR p.N173S mutation is a loss-of-function variant that fails to rescue IBD in intestinal Adar deficient mice, whereas JAK1/2 inhibitor Ruxolitinib attenuates IBD. We conclude that the ADAR-dsRNA/ERVs-MDA5-JAK/STAT axis is a potential therapeutic target for IBD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


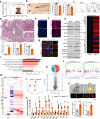
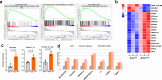
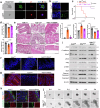

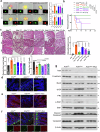
References
-
- Jairath, V. & Feagan, B. G. Global burden of inflammatory bowel disease. Lancet Gastroenterol. Hepatol.5, 2–3 (2020). - PubMed
-
- Baumgart, D. C. & Carding, S. R. Inflammatory bowel disease: cause and immunobiology. Lancet369, 1627–1640 (2007). - PubMed
-
- Na, Y. R., Stakenborg, M., Seok, S. H. & Matteoli, G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol.16, 531–543 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- DK135538/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- ES030429/U.S. Department of Health & Human Services | NIH | National Institute of Environmental Health Sciences (NIEHS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

