Prolonged glucagon exposure rewires lipid oxidation and drives diabetic kidney disease progression
- PMID: 41022721
- PMCID: PMC12479795
- DOI: 10.1038/s41467-025-63529-5
Prolonged glucagon exposure rewires lipid oxidation and drives diabetic kidney disease progression
Abstract
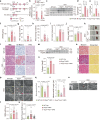
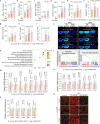
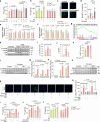
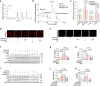
Diabetic kidney disease (DKD) is the leading cause of end-stage kidney disease. Tubular abnormalities may precede glomerular pathology and indicate functional progression of DKD. Here, we find glucagon injection exacerbates lipid accumulation and renal injury, in addition to causing morphological changes in proximal tubules, podocytes, and mitochondria in the early phase of DKD in mice. However, the specific knockdown or knockout of Gcgr in renal tubular epithelial cells almost completely halts DKD development. In contrast to the effect of short-term glucagon stimulation, long-term glucagon exposure leads to the reversal of glucagon action (glucagon reversal) in proximal tubular epithelial cells (PTECs), which is characterized by reduced energy production and an increase in lipogenesis through Gcgr-PKA-Creb-mTORC1 pathway. Accordingly, anti-GCGR antibody treatment strongly blocks the pathogenesis of DKD induced by both type 2 and type 1 diabetes. Thus, our results highlight a previously unrecognized role of glucagon/Gcgr signaling in PTEC lipogenesis and DKD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Hai Yan is an employee in and holding shares of REMD Biotherapeutics stock.
Figures



References
-
- Alicic, R. & Nicholas, S. B. Diabetic kidney disease back in focus: management field guide for health care professionals in the 21st century. Mayo Clin. Proc.97, 1904–1919 (2022). - PubMed
-
- Groth, T. et al. Wearable and implantable artificial kidney devices for end-stage kidney disease treatment: current status and review. Artif. Organs47, 649–666 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

