In vivo CRISPR screening in head and neck cancer reveals Uchl5 as an immunotherapy target
- PMID: 41022734
- PMCID: PMC12480505
- DOI: 10.1038/s41467-025-63592-y
In vivo CRISPR screening in head and neck cancer reveals Uchl5 as an immunotherapy target
Abstract
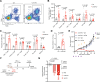
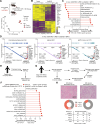
Recurrent/metastatic head and neck squamous cell carcinoma (HNSCC) is an aggressive malignancy with a significant unmet need for enhancing immunotherapy response given current modest efficacy. Here, we perform an in vivo CRISPR screen in an HNSCC mouse model to identify immune evasion genes. We identify several regulators of immune checkpoint blockade (ICB) response, including the ubiquitin C-terminal hydrolase 5 (UCHL5). Loss of Uchl5 in tumors increases CD8+ T cell infiltration and improved ICB responses. Uchl5 deficiency attenuates extracellular matrix (ECM) production and epithelial-mesenchymal-transition (EMT) transcriptional programs, which contribute to stromal desmoplasia, a histologic finding we describe as associated with reduced anti-PD1 response in human HNSCCs. COL17A1, a collagen highly and specifically expressed in HNSCC, mediates in part Uchl5-mediated immune evasion. Our findings suggest an unappreciated role for UCHL5 in promoting EMT in HNSCC and highlight ECM modulation as a strategy to improve immunotherapy responses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.T.M. receives research funding from Calico Life Sciences, LLC. R.T.M. has received speaking or consulting fees from Bristol Myers Squibb, Gilead Sciences, Kumquat Biosciences, Immunai Therapeutics, and BioNTech. R.U. reports grants and personal fees from Merck, personal fees from Regeneron, and Daichi-Sankyo. The MOC models developed by R.U. have been filed with the Washington University Office of Technology Management and are licensed for distribution by Kerafast. The remaining authors declare no competing interests.
Figures



References
-
- Ferlay, J. et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer144, 1941–1953 (2019). - PubMed
-
- Villanueva, L., Álvarez-Errico, D. & Esteller, M. The contribution of epigenetics to cancer immunotherapy. Trends Immunol.41, 676–691 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

