TP53 variant clusters stratify phenotypic diversity in germline carriers and reveal an osteosarcoma-prone subgroup
- PMID: 41022739
- PMCID: PMC12480935
- DOI: 10.1038/s41467-025-63528-6
TP53 variant clusters stratify phenotypic diversity in germline carriers and reveal an osteosarcoma-prone subgroup
Abstract
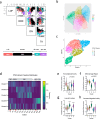
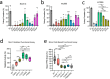
Li-Fraumeni syndrome (LFS) has recently been redefined as a 'spectrum' cancer predisposition disorder to reflect its broad phenotypic heterogeneity. This variability is thought to stem in part from the diverse functional impacts of TP53 variants, although the underlying mechanisms remain poorly understood and there is an unmet clinical need for effective risk stratification. Here, we apply unsupervised clustering to functional datasets and identify distinct TP53 variant groups with clinical relevance, including a monomeric subgroup enriched in osteosarcoma cases. In cellular validation assays, dermal fibroblasts from carriers of more functionally impaired variants exhibit increased metabolic growth rates, mirroring trends observed in cluster-stratified clinical outcomes. These findings demonstrate the feasibility of developing diagnostic assays to guide personalized cancer risk assessment. More broadly, our results show that nuances in TP53 dysfunction shape the germline TP53-related cancer susceptibility spectrum and provide a framework for functionally delineating variant carriers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Li, F. P. & Fraumeni, J. F. Jr. Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J. Natl Cancer Inst.43, 1365–1373 (1969). - PubMed
-
- Malkin, D. et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science250, 1233–1238 (1990). - PubMed
-
- Srivastava, S. et al. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature348, 747–749 (1990). - PubMed
-
- Birch, J. M. et al. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res54, 1298–1304 (1994). - PubMed
-
- Eeles, R. A. Germline mutations in the TP53 gene. Cancer Surv.25, 101–124 (1995). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

