The pericardium forms as a distinct structure during heart formation
- PMID: 41022776
- PMCID: PMC12480573
- DOI: 10.1038/s41467-025-63599-5
The pericardium forms as a distinct structure during heart formation
Abstract
The heart is formed from diverse cell lineages that assemble into a functional unit, including the pericardium, a mesothelial sac that supports movement, homeostasis, and immune responses. However, its developmental origins remain unresolved. Here, we find the pericardium forms within the lateral plate mesoderm from dedicated mesothelial progenitors that are distinct from the classic heart field. Imaging of transgenic zebrafish reporters documents lateral plate mesoderm cells that emerge lateral of the heart field among a continuous mesothelial progenitor band. Single-cell transcriptomics and trajectories of hand2-expressing lateral plate mesoderm reveal distinct populations of mesothelial precursors, including pericardial precursors. Their mesothelial gene expression signature is conserved in mammals and carries over to post-natal development. Light sheet imaging and machine learning-supported cell tracking documents the migration of pericardial precursors from the edge of the heart field to form the pericardial cavity. Genetic perturbations reveal this process occurs independently of heart formation, with canonical Wnt/β-catenin signaling modulating pericardial cell number and tissue rigidity. We connect the pathological expression of secreted Wnt antagonists of the SFRP family found in pediatric dilated cardiomyopathy to increased pericardial stiffness in neonatal rats. Altogether, our data integrate pericardium formation as an independent process into heart morphogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
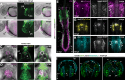

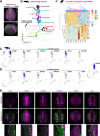

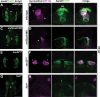
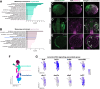
Update of
-
The pericardium forms as a distinct structure during heart formation.bioRxiv [Preprint]. 2024 Sep 28:2024.09.18.613484. doi: 10.1101/2024.09.18.613484. bioRxiv. 2024. Update in: Nat Commun. 2025 Sep 29;16(1):8566. doi: 10.1038/s41467-025-63599-5. PMID: 39345600 Free PMC article. Updated. Preprint.
References
-
- Jaworska-Wilczynska, M., Trzaskoma, P., Szczepankiewicz, A. A. & Hryniewiecki, T. Pericardium: structure and function in health and disease. Folia Histochem. Cytobiol.54, 121–125 (2016). - PubMed
-
- Schulte, I., Schlueter, J., Abu-Issa, R., Brand, T. & Männer, J. Morphological and molecular left-right asymmetries in the development of the proepicardium: a comparative analysis on mouse and chick embryos. Dev. Dyn.236, 684–695 (2007). - PubMed
-
- Ratajska, A., Czarnowska, E. & Ciszek, B. Embryonic development of the proepicardium and coronary vessels. Int. J. Dev. Biol.52, 229–236 (2008). - PubMed
-
- Kruithof, B. P. T. et al. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev. Biol.295, 507–522 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- R21 AG080257/AG/NIA NIH HHS/United States
- 24DIVSUP1281949/AHA/American Heart Association-American Stroke Association/United States
- 5K24HL150630-02/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- K24 HL150630/HL/NHLBI NIH HHS/United States
- K25 HL148386/HL/NHLBI NIH HHS/United States
- 1R01HL168097-01A1/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- T32 GM141742/GM/NIGMS NIH HHS/United States
- R01 HL169578/HL/NHLBI NIH HHS/United States
- R01 AA031043/AA/NIAAA NIH HHS/United States
- R01 DK129350/DK/NIDDK NIH HHS/United States
- R01 HL168097/HL/NHLBI NIH HHS/United States
- F31 HL167580/HL/NHLBI NIH HHS/United States
- 1R01DK129350-01A1/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
LinkOut - more resources
Full Text Sources

