Uncertainty-aware ensemble of foundation models differentiates glioblastoma from its mimics
- PMID: 41022881
- PMCID: PMC12480093
- DOI: 10.1038/s41467-025-64249-6
Uncertainty-aware ensemble of foundation models differentiates glioblastoma from its mimics
Abstract
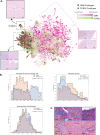
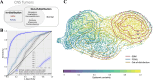
Accurate pathological diagnosis is crucial in guiding personalized treatments for patients with central nervous system cancers. Distinguishing glioblastoma and primary central nervous system lymphoma is particularly challenging due to their overlapping pathology features, despite the distinct treatments required. To address this challenge, we establish the Pathology Image Characterization Tool with Uncertainty-aware Rapid Evaluations (PICTURE) system using 2141 pathology slides collected worldwide. PICTURE employs Bayesian inference, deep ensemble, and normalizing flow to account for the uncertainties in its predictions and training set labels. PICTURE accurately diagnoses glioblastoma and primary central nervous system lymphoma with an area under the receiver operating characteristic curve (AUROC) of 0.989, with the results validated in five independent cohorts (AUROC = 0.924-0.996). In addition, PICTURE identifies samples belonging to 67 types of rare central nervous system cancers that are neither gliomas nor lymphomas. Our approaches provide a generalizable framework for differentiating pathological mimics and enable rapid diagnoses for central nervous system cancer patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.-H.Y. is an inventor of U.S. Patent 10,832,406. This patent is assigned to Harvard University and is not directly related to this manuscript. K.-H.Y. was a consultant for Curatio.DL (not related to this work). All other authors have nothing to disclose.
Figures



References
MeSH terms
Grants and funding
- R35GM142879/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- HT9425-23-1-0523/U.S. Department of Defense (United States Department of Defense)
- RSG-24-1253761-01-ESED/American Cancer Society (American Cancer Society, Inc.)
- Google Research Scholar Award/Google
- Dean's Innovation Award/Harvard Medical School
LinkOut - more resources
Full Text Sources
Medical

