Elastin-derived extracellular matrix fragments drive aging through innate immune activation
- PMID: 41023316
- PMCID: PMC12705442
- DOI: 10.1038/s43587-025-00961-8
Elastin-derived extracellular matrix fragments drive aging through innate immune activation
Abstract
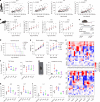
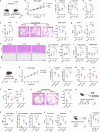
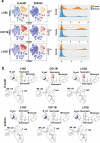
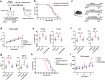
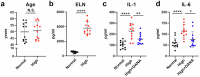
The roles of cells in systemic aging have been systematically investigated, while the roles of the extracellular matrix (ECM) and its degradation have been largely overlooked. Herein, we show that the serum contents of elastin-, hyaluronic acid- and fibronectin-derived fragments are all positively correlated with age. Elastin-derived fragments exhibited the most potent lifespan-shortening effects in mice and a positive correlation with various aging indicators in a human cohort (n = 1,068). Mechanistically, the VGVAPG oligopeptide (E-motif) in elastin-derived fragments activated monocytes and macrophages through NEU1, a component of the elastin receptor complex, which consequently caused an inflammatory response. Therapeutically, a NEU1 inhibitor extended lifespan by up to 17% in wild-type naturally aged mice and alleviated aging-related phenotypes in wild-type mice, immune-humanized mice and pigs. This study uncovers that degraded ECM acts as a circulating driver of aging, providing an anti-aging intervention strategy focused on particular elastin fragment signals.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Selman, M. & Pardo, A. Fibroageing: an ageing pathological feature driven by dysregulated extracellular matrix–cell mechanobiology. Ageing Res. Rev.70, 101393 (2021). - PubMed
-
- López-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell186, 243–278 (2023). - PubMed
-
- Smith, E. R. et al. Elastin degradation is associated with progressive aortic stiffening and all-cause mortality in predialysis chronic kidney disease. Hypertension59, 973–978 (2012). - PubMed
-
- Petersen, E., Wågberg, F. & Angquist, K.-A. Serum concentrations of elastin-derived peptides in patients with specific manifestations of atherosclerotic disease. Eur. J. Vasc. Endovasc. Surg.24, 440–444 (2002). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

