Uncovering hidden protein modifications with native top-down mass spectrometry
- PMID: 41023435
- PMCID: PMC12510877
- DOI: 10.1038/s41592-025-02846-5
Uncovering hidden protein modifications with native top-down mass spectrometry
Abstract
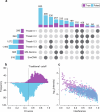
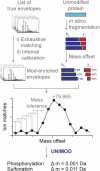
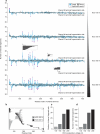
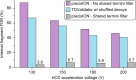
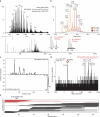
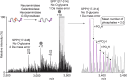
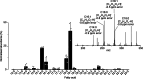
Protein modifications drive dynamic cellular processes by modulating biomolecular interactions, yet capturing these modifications within their native structural context remains a significant challenge. Native top-down mass spectrometry promises to preserve the critical link between modifications and interactions. However, current methods often fail to detect uncharacterized or low-abundance modifications, limiting insights into proteoform diversity. To address this gap, we introduce precise and accurate Identification Of Native proteoforms (precisION), an interactive end-to-end software package that leverages a robust, data-driven fragment-level open search to detect, localize and quantify 'hidden' modifications within intact protein complexes. Applying precisION to four therapeutically relevant targets-PDE6, ACE2, osteopontin (SPP1) and a GABA transporter (GAT1)-we discover undocumented phosphorylation, glycosylation and lipidation, and resolve previously uninterpretable density in an electron cryo-microscopy map of GAT1. As an open-source software package, precisION offers an intuitive means for interpreting complex protein fragmentation data. This tool will empower the community to unlock the potential of native top-down mass spectrometry, advancing integrative structural biology, molecular pathology and drug development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.L.B., T.J.E., C.A.L. and C.V.R. are listed as inventors on a pending European patent application EP24190466 entitled ‘Improved mass spectrometry methods’, assigned to the University of Oxford, describing approaches for analyzing top-down mass spectra. C.V.R. is a cofounder of and scientific advisor at OMass Therapeutics. The other authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

