Benchmarking multi-slice integration and downstream applications in spatial transcriptomics data analysis
- PMID: 41024097
- PMCID: PMC12477787
- DOI: 10.1186/s13059-025-03796-z
Benchmarking multi-slice integration and downstream applications in spatial transcriptomics data analysis
Abstract
Background: Spatial transcriptomics preserves spatial context of tissues while capturing gene expression. As the technology advances, researchers are increasingly generating data from multiple tissue sections, creating a growing demand for multi-slice integration methods. These methods aim to generate spatially aware embeddings that jointly capture spatial and transcriptomic information, preserving biological signals while mitigating technical artifacts such as batch effects. However, the reliability of these methods varies, and the growing diversity of technologies makes integration even more challenging. This underscores the need for a comprehensive benchmark to evaluate their performance, which is still lacking.
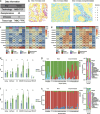
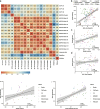
Results: To systematically evaluate the performance of multi-slice integration methods, we propose a comprehensive benchmarking framework covering four key tasks that form an upstream-to-downstream pipeline: multi-slice integration, spatial clustering, spatial alignment, slice representation. For each task, we perform detailed analyses of the methods and provide actionable recommendations. Our results reveal substantial data-dependent variation in performance across tasks. We further investigate the relationships between upstream and downstream tasks, showing that downstream performance often depends on upstream quality.
Conclusions: Our study provides a comprehensive benchmark of 12 multi-slice integration methods across four key tasks using 19 diverse datasets. Our results reveal that method performance is highly dependent on application context, dataset size, and technology. We also identified strong interdependencies between upstream and downstream tasks, highlighting the importance of robust early-stage analysis.
Keywords: Spatial multi-slice integration; Spatial transcriptomics; Systematic benchmark.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Moses L, Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022. 10.1038/s41592-022-01409-2. - PubMed
-
- Khaliq AM, et al. Spatial transcriptomic analysis of primary and metastatic pancreatic cancers highlights tumor microenvironmental heterogeneity. Nat Genet. 2024;56:2455–65. 10.1038/s41588-024-01914-4. - PubMed
MeSH terms
Grants and funding
- 324B2013/National Natural Science Foundation of China
- 62303119/National Natural Science Foundation of China
- 32341008/National Natural Science Foundation of China
- 23YF1403000/Shanghai Science and Technology Development Foundation
- 22CGA02/Chenguang Program of Shanghai Education Development Foundation and Shanghai Municipal Education Commission
LinkOut - more resources
Full Text Sources
Research Materials

