A drug repurposing approach reveals targetable epigenetic pathways in Plasmodium vivax hypnozoites
- PMID: 41025349
- PMCID: PMC12483515
- DOI: 10.7554/eLife.98221
A drug repurposing approach reveals targetable epigenetic pathways in Plasmodium vivax hypnozoites
Abstract
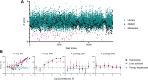
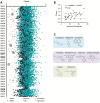
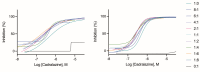
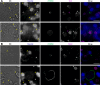
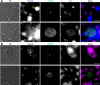
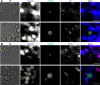
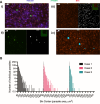
Radical cure of Plasmodium vivax malaria must include elimination of quiescent 'hypnozoite' forms in the liver; however, the only FDA-approved treatments are contraindicated in many vulnerable populations. To identify new drugs and drug targets for hypnozoites, we screened the Repurposing, Focused Rescue, and Accelerated Medchem (ReFRAME) library and a collection of epigenetic inhibitors against P. vivax liver stages. From both libraries, we identified inhibitors targeting epigenetics pathways as selectively active against P. vivax and P. cynomolgi hypnozoites. These include DNA methyltransferase inhibitors as well as several inhibitors targeting histone post-translational modifications. Immunofluorescence staining of Plasmodium liver forms showed strong nuclear 5-methylcystosine signal, indicating liver stage parasite DNA is methylated. Using bisulfite sequencing, we mapped genomic DNA methylation in sporozoites, revealing DNA methylation signals in most coding genes. We also demonstrated that methylation level in proximal promoter regions as well as in the first exon of the genes may affect, at least partially, gene expression in P. vivax. The importance of selective inhibitors targeting epigenetic features on hypnozoites was validated using MMV019721, an acetyl-CoA synthetase inhibitor that affects histone acetylation and was previously reported as active against P. falciparum blood stages. In summary, our data indicate that several epigenetic mechanisms are likely modulating hypnozoite formation or persistence and provide an avenue for the discovery and development of improved radical cure antimalarials.
Keywords: DNA methylation; P. cynomolgi; P. falciparum; P. vivax; Plasmodium cynomolgi; Plasmodium vivax; biochemistry; chemical biology; hypnozoites; infectious disease; malaria; microbiology; primary hepatocytes; rhesus macaque.
© 2024, Maher et al.
Conflict of interest statement
SM, MB, AV, CA, MG, YA, MA, MC, AC, AC, WC, CC, MG, HJ, SJ, TL, SL, JM, AO, VP, KP, JP, JP, CR, AR, SS, CS, JS, SS, SS, RU, YW, PW, JY, JP, CM, CJ, FN, BW, KL, DK No competing interests declared, EF, AH, SM AH-C, VC, ELF, and SAM are employees of the Novartis Institute for Tropical Disease, BC BC is an employee of MMV, VC AH-C, VC, ELF, and SAM are employees of the Novartis Institute for Tropical Disease,, KC, TM TM and KC are employees of BioIVT
Figures


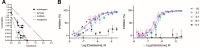









Update of
-
A Drug Repurposing Approach Reveals Targetable Epigenetic Pathways in Plasmodium vivax Hypnozoites.bioRxiv [Preprint]. 2024 Mar 25:2023.01.31.526483. doi: 10.1101/2023.01.31.526483. bioRxiv. 2024. Update in: Elife. 2025 Sep 30;13:RP98221. doi: 10.7554/eLife.98221. PMID: 36778461 Free PMC article. Updated. Preprint.
References
-
- Ansari HR, Templeton TJ, Subudhi AK, Ramaprasad A, Tang J, Lu F, Naeem R, Hashish Y, Oguike MC, Benavente ED, Clark TG, Sutherland CJ, Barnwell JW, Culleton R, Cao J, Pain A. Genome-scale comparison of expanded gene families in Plasmodium ovale wallikeri and Plasmodium ovale curtisi with Plasmodium malariae and with other Plasmodium species. International Journal for Parasitology. 2016;46:685–696. doi: 10.1016/j.ijpara.2016.05.009. - DOI - PubMed
-
- Antonova-Koch Y, Meister S, Abraham M, Luth MR, Ottilie S, Lukens AK, Sakata-Kato T, Vanaerschot M, Owen E, Jado JC, Maher SP, Calla J, Plouffe D, Zhong Y, Chen K, Chaumeau V, Conway AJ, McNamara CW, Ibanez M, Gagaring K, Serrano FN, Eribez K, Taggard CM, Cheung AL, Lincoln C, Ambachew B, Rouillier M, Siegel D, Nosten F, Kyle DE, Gamo FJ, Zhou Y, Llinás M, Fidock DA, Wirth DF, Burrows J, Campo B, Winzeler EA. Open-source discovery of chemical leads for next-generation chemoprotective antimalarials. Science. 2018;362:eaat9446. doi: 10.1126/science.aat9446. - DOI - PMC - PubMed
-
- Avery VM, Bashyam S, Burrows JN, Duffy S, Papadatos G, Puthukkuti S, Sambandan Y, Singh S, Spangenberg T, Waterson D, Willis P. Screening and hit evaluation of a chemical library against blood-stage Plasmodium falciparum. Malaria Journal. 2014;13:190. doi: 10.1186/1475-2875-13-190. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- RD/17/0042/Medicines for Malaria Venture
- R01 AI136511/AI/NIAID NIH HHS/United States
- INV-031788/GATES/Gates Foundation/United States
- #NIFA-Hatch-225935/University of California, Riverside
- HHSN272201200031C/AI/NIAID NIH HHS/United States
- #OPP1107194/Bill and Melinda Gates Foundation
- #OPP1023601/Bill and Melinda Gates Foundation
- WT_/Wellcome Trust/United Kingdom
- RD/15/0022/Medicines for Malaria Venture
- #HHSN272201200031C/NH/NIH HHS/United States
- S10 OD026929/OD/NIH HHS/United States
- 1R01 AI136511/NH/NIH HHS/United States
- INV-031788/GATES/Gates Foundation/United States
LinkOut - more resources
Full Text Sources

