Protein corona formed on lipid nanoparticles compromises delivery efficiency of mRNA cargo
- PMID: 41027853
- PMCID: PMC12485112
- DOI: 10.1038/s41467-025-63726-2
Protein corona formed on lipid nanoparticles compromises delivery efficiency of mRNA cargo
Abstract
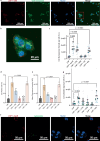
Lipid nanoparticles (LNPs) are the most clinically advanced nonviral RNA-delivery vehicles, though challenges remain in fully understanding how LNPs interact with biological systems. In vivo, proteins form an associated corona on LNPs that redefines their physicochemical properties and influences delivery outcomes. Despite its importance, the LNP protein corona is challenging to study owing to the technical difficulty of selectively recovering soft nanoparticles from biological samples. Herein, we develop a quantitative, label-free mass spectrometry-based proteomics approach to characterize the protein corona on LNPs. Critically, this protein corona isolation workflow avoids artifacts introduced by the presence of endogenous nanoparticles in human biofluids. We apply continuous density gradient ultracentrifugation for protein-LNP complex isolation, with mass spectrometry for protein identification normalized to protein composition in the biofluid alone. With this approach, we quantify proteins consistently enriched in the LNP corona including vitronectin, C-reactive protein, and alpha-2-macroglobulin. We explore the impact of these corona proteins on cell uptake and mRNA expression in HepG2 human liver cells, and find that, surprisingly, increased levels of cell uptake do not correlate with increased mRNA expression in part due to protein corona-induced lysosomal trafficking of LNPs. Our results underscore the need to consider the protein corona in the design of LNP-based therapeutics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.A.W. is an inventor on US patents 9,227,917 (2016) and 9,439,968 (2016) related to the LNPs described here and is a consultant for several companies translating nonviral RNA delivery systems. The remaining authors declare no competing interests.
Figures




Update of
-
Protein corona formed on lipid nanoparticles compromises delivery efficiency of mRNA cargo.bioRxiv [Preprint]. 2025 Jan 24:2025.01.20.633942. doi: 10.1101/2025.01.20.633942. bioRxiv. 2025. Update in: Nat Commun. 2025 Sep 30;16(1):8699. doi: 10.1038/s41467-025-63726-2. PMID: 39896592 Free PMC article. Updated. Preprint.
References
-
- Gilleron, J. et al. Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol.31, 638–646 (2013). - PubMed
-
- Hajj, K. A. & Whitehead, K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater.2, 1–17 (2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

