The critical human pathogen Acinetobacter baumannii exhibits light-regulated circadian rhythms
- PMID: 41028287
- PMCID: PMC12484797
- DOI: 10.1038/s42003-025-08732-2
The critical human pathogen Acinetobacter baumannii exhibits light-regulated circadian rhythms
Abstract
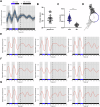
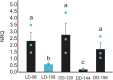
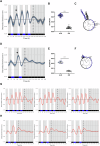
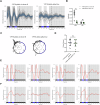
Acinetobacter baumannii is recognized as the paradigm of multidrug-resistant superbug, topping the WHO priority list of critical human pathogens. Interestingly, it senses and responds to blue light, which modulates global aspects of its physiology, including pathogenicity. We hypothesized that light could serve as a signal to synchronize the bacterial physiology to the host's behavior and/or to the environment. At environmental temperatures, light regulation is mainly governed by the photoreceptor BlsA. In this work, we identified the existence of daily rhythms in blsA promoter activity displaying a robust response to light, as well as endogenous circadian rhythms in A. baumannii. In fact, we show that blsA promoter activity can be synchronized to 24-hour blue light-dark cycles, which immediately resynchronizes after a phase shift. Upon release to constant darkness, bacterial populations present free-running oscillations with a period close to 24 hours. Furthermore, our data indicate that BlsA is involved in synchronization to light-dark cycles. In fact, under constant darkness without previous entrainment, A. baumannii is rhythmic, but acrophases are not clusterized, behaving as the blsA mutant under light-dark cycles. Our work contributes to the developing field of circadian clocks in bacterial pathogens, which could impact in the microorganisms' lifestyle and pathogenicity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

