TP63 mediates the generation of tumour-specific chromatin loops that underlie MYC activation in radiation-induced tumorigenesis
- PMID: 41028778
- PMCID: PMC12484796
- DOI: 10.1038/s41467-025-63754-y
TP63 mediates the generation of tumour-specific chromatin loops that underlie MYC activation in radiation-induced tumorigenesis
Abstract
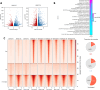
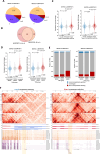
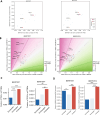
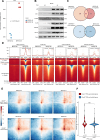
Alterations in 3D chromatin conformation may disrupt the interplay between promoters and distal enhancers. How gene regulatory circuits are reshaped during ionizing radiation-induced tumorigenesis remains unclear, and little is known about the mediators that drive these processes. To decipher the chromatin alterations in radiation-induced lung cancer, we performed ATAC-seq, RNA-seq and Hi-C analyses of human bronchial epithelial cells and corresponding radiation-induced malignantly transformed cell lines. We found that this malignant transformation is accompanied by chromatin switching from the inactive B compartment to the active A compartment, an increased number of TADs and gained ATAC-seq peaks that mediate new distal chromatin contacts. We identified tumour protein 63 (TP63) as a mediator of new chromatin-accessible sites that anchor tumour-specific chromatin contacts in radiation-induced tumour cells. A TP63-mediated accessible chromatin site anchors a tumour-specific TAD boundary and multiple tumour-specific chromatin loops, which might underlie MYC oncogene activation during malignant transformation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Chatterjee, A., Rodger, E. J. & Eccles, M. R. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin. Cancer Biol.51, 149–159 (2018). - PubMed
-
- Zhu, H. et al. Candidate cancer driver mutations in distal regulatory elements and long-range chromatin interaction networks. Mol. Cell77, 1307–1321.e10 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

