CRISPR/Cas9-mediated mutation of GhCAD decreases the gossypol content of cottonseed
- PMID: 41029731
- PMCID: PMC12486840
- DOI: 10.1186/s13036-025-00556-2
CRISPR/Cas9-mediated mutation of GhCAD decreases the gossypol content of cottonseed
Abstract
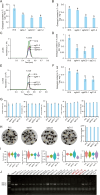
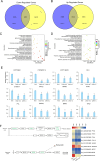
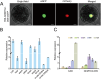
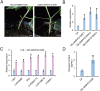
Cottonseed is the most important byproduct of cotton production. However, high free gossypol contents limit the application of cottonseed in the food or feed industry. In this study, CRISPR/Cas9 technology was used to knock out the (+)-δ-cadinene synthase gene (GhCAD) to decrease the gossypol content. Gossypol levels decreased approximately 64% in cottonseeds and leaves following the targeted mutation of GhCAD. If only GhCAD1-A was edited, the seed gossypol content decreased by approximately 46%, but there were no major changes in the leaf gossypol content. In addition, the protein and fatty acid (C16:0, C18:1, and C18:2) profiles of the transgenic cotton seeds were similar to those of the control cotton seeds. Furthermore, transcriptome analysis revealed that the jasmonic acid signal transduction pathway was significantly enriched among the DEGs, and GhMYC2-D09 expression was down-regulated. Silencing of GhMYC2-D09 via virus-induced gene silencing decreased the expression of gossypol biosynthesis-related genes, ultimately restricting the accumulation of gossypol in cotton leaves. In contrast, the overexpression of GhMYC2-D09 in hairy roots had the opposite effect. Dual-luciferase assays revealed that GhMYC2-D09 can activate the expression of GhCAD1-A and GhCAD1-C, but Y1H assays revealed that GhMYC2-D09 cannot bind directly to GhCAD promoters. In conclusion, we used CRISPR/Cas9 technology to silence GhCAD expression and developed new genetic resources for generating low-gossypol cotton materials. Furthermore, we characterized GhMYC2-D09 as a transcription factor that increases gossypol biosynthesis. These findings may provide new insights to further elucidate the regulatory network of gossypol biosynthesis.
Keywords: GhCAD; CRISPR/cas9; Cotton; Gossypol.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Rogers GM, Poore MH, Paschal JC. Feeding cotton products to cattle. Veterinary Clin North Am Food Anim Pract. 2002;18(2):267–94. - PubMed
-
- Dowd MK, Boykin DL, Meredith WR Jr, Todd Campbell B, Bourland FM, Gannaway JR, Glass KM, Zhang J. Fatty acid profiles of cottonseed genotypes from the National cotton variety trials. J Cotton Sci. 2010;14(2):64–73.
-
- Yuan X, Nagamine R, Tanaka Y, Tsai WT, Jiang Z, Takeyama A, Imaizumi K, Sato M. The effects of dietary Linoleic acid on reducing serum cholesterol and atherosclerosis development are nullified by a high-cholesterol diet in male and female apoE-deficient mice. Br J Nutr. 2023;129(5):737–44. - PubMed
-
- Shahid LA, Saeed MA, Amjad N. Present status and future prospects of mechanized production of oilseed crops in Pakistan-a review. Pakistan J Agric Res. 2010;23:83–93.
-
- Zia MA, Shah SH, Shoukat S, Hussain Z, Khan SU, Shafqat N. Physicochemical features, functional characteristics, and health benefits of cottonseed oil: a review. Braz J Biol. 2021;82:e243511. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

