This is a preprint.
A sequence-specific RNA acetylation catalyst
- PMID: 41030981
- PMCID: PMC12478401
- DOI: 10.1101/2024.10.25.620255
A sequence-specific RNA acetylation catalyst
Update in
-
A sequence-specific RNA acetylation catalyst.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf217. doi: 10.1093/nar/gkaf217. Nucleic Acids Res. 2025. PMID: 40119730 Free PMC article.
Abstract
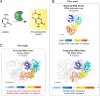
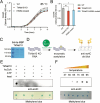
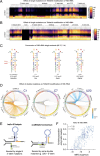
N4-acetylcytidine (ac4C) is a ubiquitous RNA modification incorporated by cytidine acetyltransferase enzymes. Here, we report the biochemical characterization of Thermococcus kodakarensis Nat10 (TkNat10), an RNA acetyltransferase involved in archaeal thermotolerance. We demonstrate that TkNat10's catalytic activity is critical for T. kodakarensis fitness at elevated temperatures. Unlike eukaryotic homologs, TkNat10 exhibits robust stand-alone activity, modifying diverse RNA substrates in a temperature, ATP, and acetyl-CoA-dependent manner. Transcriptome-wide analysis reveals TkNat10 preferentially modifies unstructured RNAs containing a 5'-CCG-3' consensus sequence. Using a high-throughput mutagenesis approach, we define sequence and structural determinants of TkNat10 substrate recognition. We find TkNat10 can be engineered to use non-native acyl-CoA donors, providing insight into its cofactor specificity. Finally, we demonstrate TkNat10's utility for site-specific acetylation of RNA oligonucleotides, enabling analysis of ac4C-dependent RNA-protein interactions. Our findings establish a framework for understanding archaeal RNA acetylation and a new tool for studying the functional consequences of ac4C in diverse RNA contexts.
Figures



References
-
- Stern L.; Schulman L. H. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J Biol Chem 1978, 253 (17), 6132–6139. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
