This is a preprint.
Benchmarking large language models for biomedical natural language processing applications and recommendations
- PMID: 41031069
- PMCID: PMC12478438
Benchmarking large language models for biomedical natural language processing applications and recommendations
Update in
-
Benchmarking large language models for biomedical natural language processing applications and recommendations.Nat Commun. 2025 Apr 6;16(1):3280. doi: 10.1038/s41467-025-56989-2. Nat Commun. 2025. PMID: 40188094 Free PMC article.
Abstract
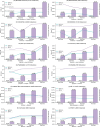
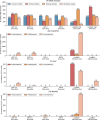
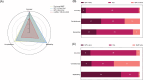
The rapid growth of biomedical literature poses challenges for manual knowledge curation and synthesis. Biomedical Natural Language Processing (BioNLP) automates the process. While Large Language Models (LLMs) have shown promise in general domains, their effectiveness in BioNLP tasks remains unclear due to limited benchmarks and practical guidelines. We perform a systematic evaluation of four LLMs-GPT and LLaMA representatives-on 12 BioNLP benchmarks across six applications. We compare their zero-shot, few-shot, and fine-tuning performance with traditional fine-tuning of BERT or BART models. We examine inconsistencies, missing information, hallucinations, and perform cost analysis. Here we show that traditional fine-tuning outperforms zero- or few-shot LLMs in most tasks. However, closed-source LLMs like GPT-4 excel in reasoning-related tasks such as medical question answering. Open-source LLMs still require fine-tuning to close performance gaps. We find issues like missing information and hallucinations in LLM outputs. These results offer practical insights for applying LLMs in BioNLP.
Conflict of interest statement
Competing Interests Statement Dr. Jingcheng Du and Dr. Hua Xu have research-related financial interests at Melax Technologies Inc. The remaining authors declare no competing interests.
Figures

References
-
- Leaman R., Islamaj R., Allot A., Chen Q., Wilbur W. J., and Lu Z., “Comprehensively identifying long Covid articles with human-in-the-loop machine learning,” Patterns, vol. 4, no. 1, 2023.
-
- Chen Q., Lee K., Yan S., Kim S., Wei C.-H., and Lu Z., “BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale,” PLoS computational biology, vol. 16, no. 4, pp. e1007617, 2020.
-
- Blake C., “Beyond genes, proteins, and abstracts: Identifying scientific claims from full-text biomedical articles,” Journal of biomedical informatics, vol. 43, no. 2, pp. 173–189, 2010. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
