CobVar-a comprehensive resource of vitamin B12-associated genomic variants
- PMID: 41031637
- PMCID: PMC12485615
- DOI: 10.1093/database/baaf049
CobVar-a comprehensive resource of vitamin B12-associated genomic variants
Abstract
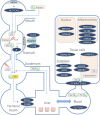
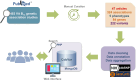
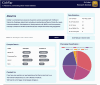
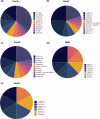
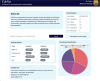
The importance of vitamin B12 (cobalamin) in numerous biological processes, including DNA synthesis and cellular energy production, underscores the need for therapeutic and public health strategies to address B12 insufficiency/deficiency in the population. Genetic variations in pathways influencing cobalamin absorption, transport, and metabolism can affect various direct and indirect measures of vitamin B12 status. To facilitate a structured approach to studying these genetic factors, we aimed to systematically curate and create a user-friendly web database that offers comprehensive data on genetic variants influencing B12 biomarkers. A PubMed search was performed for 5 B12 traits [total serum/plasma B12, holotranscobalamin (active B12), total transcobalamin, holohaptocorrin, and methylmalonic acid] resulting in 493 research publications, of which 47 relevant publications were reviewed further. The database backend was built using MongoDB and the web interface was coded in PHP, JavaScript, HTML, and CSS on an Apache HTTP server. We have manually curated and compiled the Cobalamin Associated Genetic Variant (CobVar) database, comprising a total of 324 genetic variant associations for 5 different vitamin B12 traits involving 222 unique genetic variants and 84 genes identified across several genome-wide association studies and candidate gene studies. About one-third of the total genetic variant associations have been reported in >1 independent studies and 15 variants in >1 ethnic group. FUT2 gene showed the maximum number of associations for total serum/plasma B12 (N = 39), followed by MTHFR (N = 24) and TCN2 (N = 23). The database is accessible online at https://datatools.sjri.res.in/VBG/. CobVar is a vital resource for researchers and nutritionists, offering quick access to the latest developments in B12-related genetic variant research and serves as a valuable tool for advancing personalized treatment. Database URL: https://datatools.sjri.res.in/VBG/.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
References
-
- Gibson R. Principles of Nutritional Assessment. New York: Oxford University Press, 2005. 10.1093/oso/9780195171693.001.0001 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

