SGLT2 inhibition protects kidney function by SAM-dependent epigenetic repression of inflammatory genes under metabolic stress
- PMID: 41031893
- PMCID: PMC12483609
- DOI: 10.1172/JCI188933
SGLT2 inhibition protects kidney function by SAM-dependent epigenetic repression of inflammatory genes under metabolic stress
Abstract
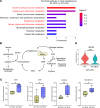
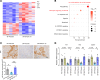
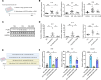
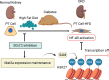
Clinically, blockade of renal glucose resorption by sodium-glucose cotransporter 2 (SGLT2) inhibitors slows progression of kidney disease, yet the underlying mechanisms are not fully understood. We hypothesized that altered renal metabolites underlie observed kidney protection when SGLT2 function is lost. S-adenosylmethionine (SAM) levels were increased in kidneys from mice lacking SGLT2 function on a diabetogenic high-fat diet (SPHFD) compared with WT mice fed HFD. Elevated SAM in SPHFD was associated with improved kidney function and decreased expression of NF-κB pathway-related genes. Injured proximal tubular cells that emerged under HFD conditions in WT mice and humans consistently showed reduction in expression of the SAM synthetase Mat2a/MAT2A, while MAT2A inhibition, which reduces SAM production, abrogated kidney protection in SPHFD mice. Histone H3 lysine 27 (H3K27) repressive trimethylation of NF-κB-related genes was increased in SPHFD, consistent with SAM's role as a methyl donor. Our data support a model whereby SGLT2 loss enhances SAM levels within the kidney, leading to epigenetic repression of inflammatory genes and kidney protection under metabolic stress.
Keywords: Diabetes; Epigenetics; Metabolism; NF-kappaB; Nephrology.
Conflict of interest statement
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

