Use of DNA Methylation Profiling as a Molecular Classification Tool for Paediatric Central Nervous System Tumours: A Middle-Income Country Population-Based Study
- PMID: 41033792
- PMCID: PMC12488389
- DOI: 10.1111/nan.70041
Use of DNA Methylation Profiling as a Molecular Classification Tool for Paediatric Central Nervous System Tumours: A Middle-Income Country Population-Based Study
Abstract
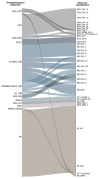
Paediatric central nervous system (CNS) tumours are the second most common childhood malignancy and the leading cause of cancer-related mortality in this age group. Histopathological diagnosis can be challenging, particularly for rare or ambiguous tumours, and may result in misclassification. To evaluate the utility of DNA methylation profiling in a middle-income country, we performed the Infinium MethylationEPIC BeadChip (Illumina) on tumours from 182 paediatric patients treated at a reference centre for paediatric oncology in Campinas, state of São Paulo, Brazil. Data were analysed using the DKFZ/Heidelberg CNS tumour classifier (v12.8). After excluding control tissue, 163 samples (89.6%) were suitable for classification; 135 (74.2%) achieved a calibrated score ≥ 0.9 and were assigned to a methylation class family. Methylation profiling resulted in a tumour subtype for 88 cases (65.7%) and changed the diagnosis in 28 cases (20.9%), identifying several rare tumour subtypes that were identified solely through methylation analysis, confirming the value of this method in improving diagnostic accuracy. This study highlights the utility of DNA methylation profiling for paediatric CNS tumours in a resource-limited setting and provides a cohort from an underrepresented middle-income population to international molecular databases.
Keywords: DNA methylation profiling; middle‐income countries; molecular classification; molecular tumour subtypes; paediatric CNS tumours.
© 2025 The Author(s). Neuropathology and Applied Neurobiology published by John Wiley & Sons Ltd on behalf of British Neuropathological Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Lam C. G., Howard S. C., Bouffet E., and Pritchard‐Jones K., “Science and Health for All Children With Cancer,” Science 363, no. 6432 (2019): 1182–1186. - PubMed
-
- Bertuccio P., Alicandro G., Malvezzi M., et al., “Childhood Cancer Mortality Trends in Europe, 1990–2017, With Focus on Geographic Differences,” Cancer Epidemiology 67 (2020): 101768. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

