Microtubule architecture connects AMOT stability to YAP/TAZ mechanotransduction and Hippo signalling
- PMID: 41034521
- PMCID: PMC12527920
- DOI: 10.1038/s41556-025-01773-z
Microtubule architecture connects AMOT stability to YAP/TAZ mechanotransduction and Hippo signalling
Abstract
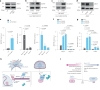
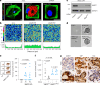
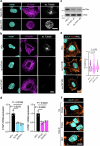
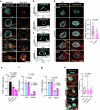
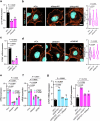
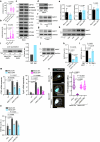
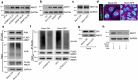
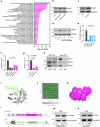
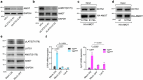
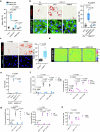
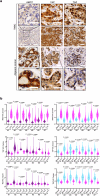
Cellular mechanotransduction is a key informational system, yet its mechanisms remain elusive. Here we unveil the role of microtubules in mechanosignalling, operating downstream of subnuclear F-actin and nuclear envelope mechanics. Upon mechanical activation, microtubules reorganize from a perinuclear cage into a radial array nucleated by centrosomes. This structural rearrangement triggers degradation of AMOT proteins, which we identify as key mechanical rheostats that sequester YAP/TAZ in the cytoplasm. AMOT is stable in mechano-OFF but degraded in mechano-ON cell states, where microtubules allow AMOT rapid transport to the pericentrosomal proteasome in complex with dynein/dynactin. This process ensures swift control of YAP/TAZ function in response to changes in cell mechanics, with experimental loss of AMOT proteins rendering cells insensitive to mechanical modulations. Ras/RTK oncogenes promote YAP/TAZ-dependent tumorigenesis by corrupting this AMOT-centred mechanical checkpoint. Notably, the Hippo pathway fine-tunes mechanotransduction: LATS kinases phosphorylate AMOT, shielding it from degradation, thereby indirectly restraining YAP/TAZ. Thus, AMOT protein stability serves as a hub linking cytoskeletal reorganization and Hippo signalling to YAP/TAZ mechanosignalling.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature474, 179–183 (2011). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

