Tracking clonal evolution during treatment in ovarian cancer using cell-free DNA
- PMID: 41034582
- PMCID: PMC12629990
- DOI: 10.1038/s41586-025-09580-0
Tracking clonal evolution during treatment in ovarian cancer using cell-free DNA
Abstract
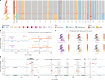
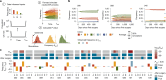
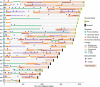
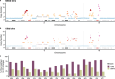
Emergence of drug resistance is the main cause of therapeutic failure in patients with high-grade serous ovarian cancer (HGSOC)1. To study drug resistance in patients, we developed CloneSeq-SV, which combines single-cell whole-genome sequencing2 with targeted deep sequencing of clone-specific genomic structural variants in time-series cell-free DNA. CloneSeq-SV exploits tumour clone-specific structural variants as highly sensitive endogenous cell-free DNA markers, enabling the relative abundance measurements and evolutionary analysis of co-existing clonal populations over the therapeutic time course. Here, using this approach, we studied 18 patients with HGSOC over a multi-year period from diagnosis to recurrence and showed that drug resistance typically arose from selective expansion of a single or small subset of clones present at diagnosis. Drug-resistant clones frequently showed interpretable and distinctive genomic features, including chromothripsis, whole-genome doubling, and high-level amplifications of oncogenes such as CCNE1, RAB25, MYC and NOTCH3. Phenotypic analysis of matched single-cell RNA sequencing data3 indicated pre-existing and clone-specific transcriptional states such as upregulation of epithelial-to-mesenchymal transition and VEGF pathways, linked to drug resistance. In one notable case, clone-specific ERBB2 amplification affected the efficacy of a secondary targeted therapy with a positive patient outcome. Together, our findings indicate that drug-resistant states in HGSOC pre-exist at diagnosis, leading to positive selection and reduced clonal complexity at relapse. We suggest these findings motivate investigation of evolution-informed adaptive treatment regimens to ablate drug resistance in future HGSOC studies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: B.W. reports grant funding by Repare Therapeutics paid to the institution, outside the submitted work, and employment of a direct family member at AstraZeneca. C.A. reports grants from Clovis, Genentech, AbbVie and AstraZeneca and personal fees from Tesaro, Eisai/Merck, Mersana Therapeutics, Roche/Genentech, AbbVie, AstraZeneca/Merck and Repare Therapeutics, outside the scope of the submitted work. C.A. reports clinical trial funding to the institution from AbbVie, AstraZeneca and Genentech/Roche; participation on a data safety monitoring board or advisory board in AstraZeneca and Merck; unpaid membership of the GOG Foundation Board of Directors and the NRG Oncology Board of Directors. M.F.B. reports consulting fees (Eli Lilly, AstraZeneca, Paige.AI), Research Support (Boundless Bio) and Intellectual Property Rights (SOPHiA Genetics). B.L. reports intellectual property rights (SOPHiA Genetics) and licensing royalties (BioLegend/Revvity). C.F. reports research funding to the institution from Merck, AstraZeneca, Genentech/Roche, Bristol Myers Squibb and Daiichi; uncompensated membership of a scientific advisory board for Merck and Genentech; and is a consultant for OncLive, Aptitude Health, Bristol Myers Squibb and Seagen, all outside the scope of this paper. D.S.C. reports membership of the medical advisory board of Verthermia Acquio and Biom’up, is a paid speaker for AstraZeneca, and holds stock of Doximity, Moderna and BioNTech. D.Z. reports institutional grants from Merck, Genentech, AstraZeneca, Plexxikon and Synthekine, and personal fees from AstraZeneca, Xencor, Memgen, Takeda, Astellas, Immunos, Tessa Therapeutics, Miltenyi and Calidi Biotherapeutics. D.Z. own a patent on use of oncolytic Newcastle Disease Virus for cancer therapy. N.R.A.-R. reports grants to the institution from Stryker/Novadaq and GRAIL, outside the submitted work. S.P.S. reports research funding from AstraZeneca and Bristol Myers Squibb, outside the scope of this work. The other authors declare no competing interests.
Figures









References
-
- Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

